Part:BBa_K3447001
Arabinose sensing system
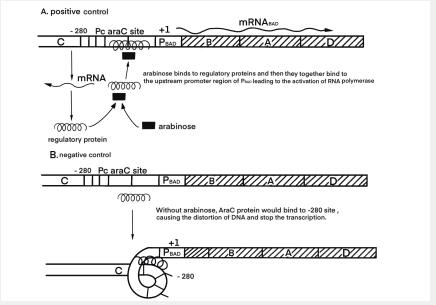
PC is a constitutive promoter that expresses the downstream araC gene. PBAD promoter is an inducible promoter, PBAD contains sequences of PC. It was verified by BBa_I0500 that the induction effect of arabinose could be realized by using only araC and PBAD.
Usage and Biology
Arabinose operon is composed of AraC protein, PBAD and PC. The araC gene translates from the PC to the left. We used the PC promoter to express AraC constitutively, and constituted the sensing part of the arabinose sensing system.
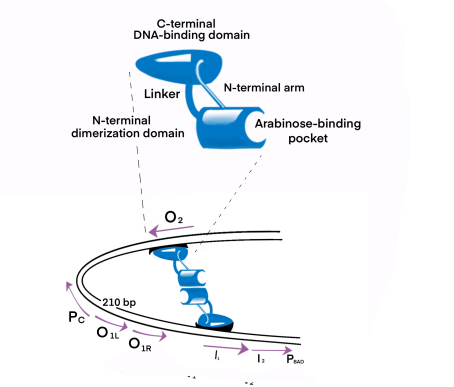
AraC gene expression product belongs to araC protein family, which is a type of two monomers with dimer structure, and each monomer with DNA binding sites of the spiral screw - corner - functional areas. It’s amino terminal determines the inducibility and it’s carboxyl terminal determines the DNA binding ability. Through this helix-rotation-helix structure, araC contacts and binds to four large sulcus regions adjacent to the DNA.
As shown in Figure 1 and 2, when arabinose is present in cells, araC, the product encoded by gene araC, binds to arabinose and binds to the upstream promoter region of PBAD to activate RNA polymerase and activate the gene after PBAD. On the contrary, when arabinose is not present, the product araC will cause DNA curling to inhibit the binding of RNA polymerase, and inhibit the expression of gene downstream of PBAD.
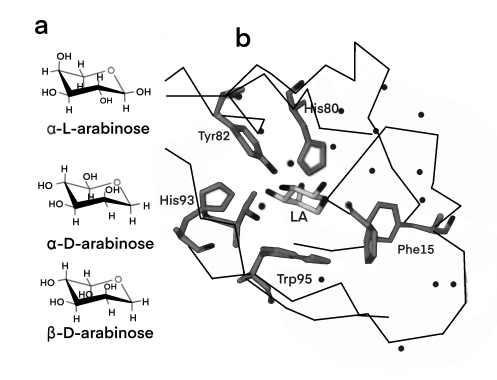
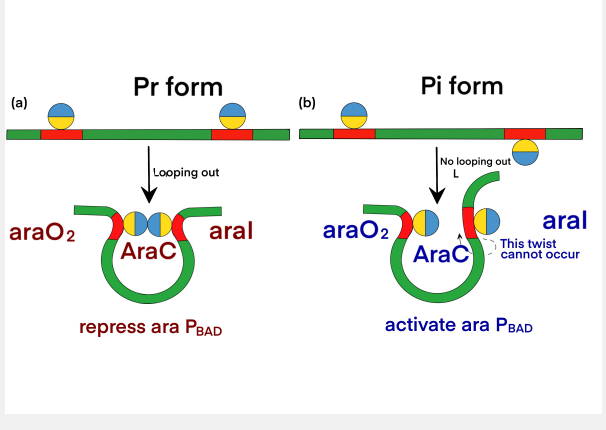
The dimerization domain is formed by folding into a pocket, combining arabinose, and dimerized by an antiparallel coiled coil. AraC stimulates RNA polymerase binding to DNA and the rate of formation of open complexes. The presence or absence of arabinose may alter the DNA contact of the upstream subunit of homodimer AraC, thereby affecting the binding of RNA polymerase. In the absence of L-arabinose, the DNA-binding domain (DBD) of the AraC dimer binds the I1 and O2 halves (separated by 210 bases) and inhibits transcription by forming a DNA ring upstream of the dimer. After PBAD binds to L-arabinose, the dimer changes the conformation, making DBD bind to the adjacent I1 and I2 halves, thus leading to transcriptional activation through interaction with RNA polymerase in PBAD. The regulatory characteristics of AraC are due to the sensitive conversion between the two conformation Pr and Pi, and the specific interaction with the inducer.
In our project, we used this part together with PC and PBAD to form the arabinose sensing system, and achieved different functions by linking different target genes downstream of PBAD.
Arabinose induces endotoxin expression: BBa_K3447104
Arabinose induces green fluorescent protein expression: BBa_K3447103
Characterization
Validation of Arabinose response system
- See details in BBa_K3447103.
This part contains an arabinose operon, which constitutes the arabinose sensing system. By connecting the sfGFP gene downstream, we were able to verify the expression intensity of this arabinose operon.
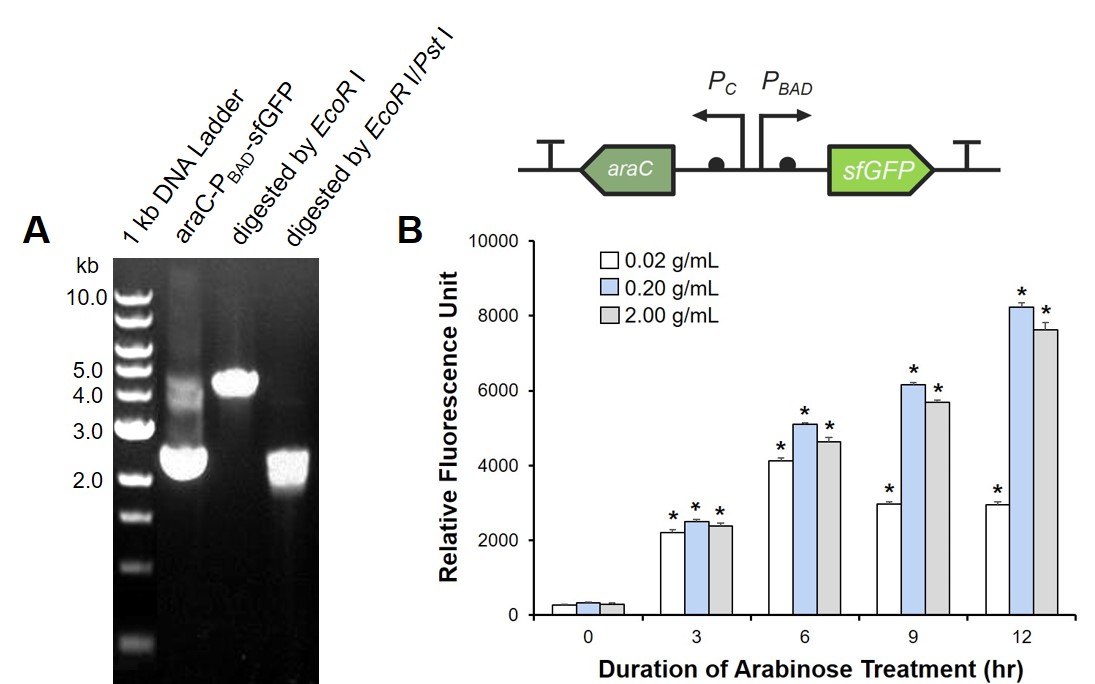
To prove the arabinose sensing system can work successfully, a sfGFP was inserted after PBAD under the induction of arabinose at a series of concentrations. As shown in Fig. 5B, the expression efficiency was highest when induced with 0.2 g/mL arabinose.
Arabinose induces endotoxin expression
- See details in BBa_K3447104.
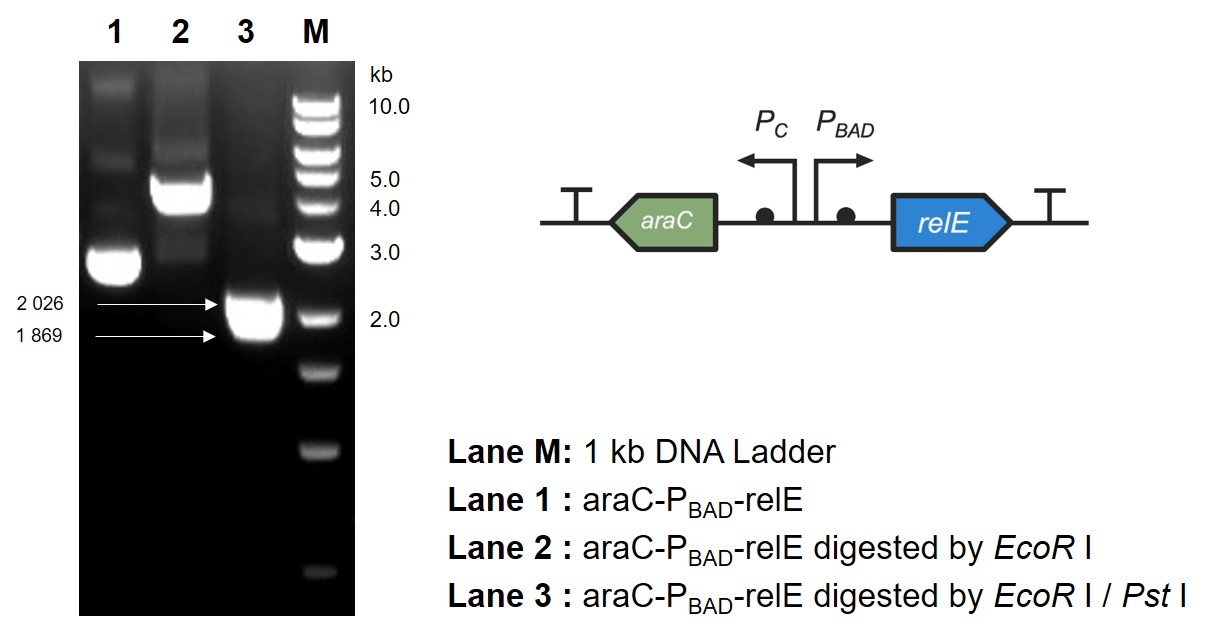
This part contains an arabinose operon, which constitutes the arabinose sensing system. By connecting the relE gene downstream, the normal growth and reproduction of bacteria will be inhibited when arabinose is present.
After the expression of arabinose operon was verified, sfGFP gene was replaced by relE gene, which causes the normal growth and reproduction of bacteria will be inhibited when there is arabinose in the medium.
The digestion and agarose gel electrophoresis are shown in Fig. 6.
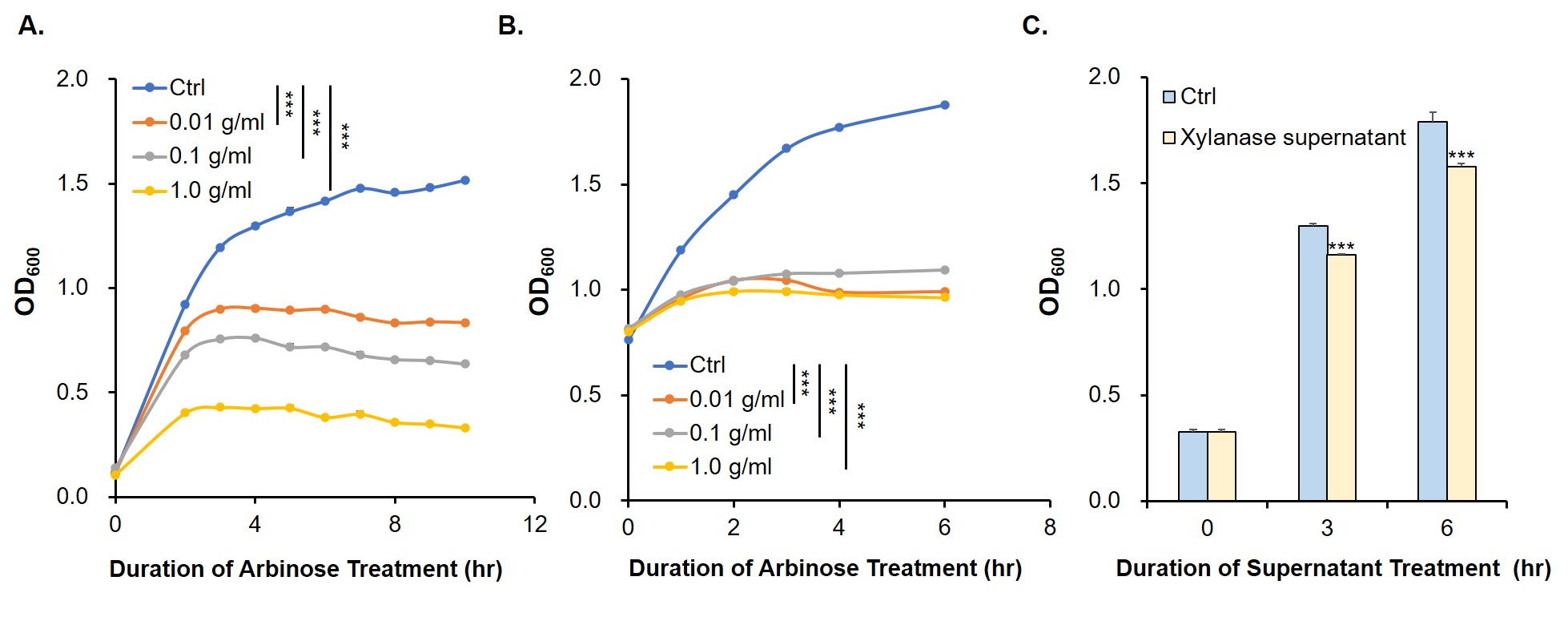
Since relE encodes a small endotoxin peptide which represses the growth of bacteria, its expression could be characterized by measuring growth curve. Fig. 7A and 7B suggest that, compared with negative control, the E. coli cells cultured with standard arabinose can be inhibited for growth and reproduction. Finally, in order to verify whether the xylanase produced by E. coli could manage to hydrolyze xylan to arabinose and, by binding to the araC, turn on the expression of relE in the downstream of PBAD, thus proving its function by measuring the growth situation (Fig. 7C).
Design
Design Notes
We added some synonymous mutations to avoid part rules.
Source
We found this sequence data in the previous iGEM teams (Glasgow 2017 and BBa_I0500) and in GenBank.
References
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1216
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1155
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 990
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1
Illegal BsaI.rc site found at 1245
Illegal SapI site found at 972
| None |