Part:BBa_K3427001
knt-gfp entangled CDSs for aminoglycoside resistance and GFP
Generation of the biobrick sequence with CAMEOS software We used a software named Cameos which can produce a sequence with entangled genes, i.e. overlapping coding sequences where the entire coding sequence of gene A is comprised within the sequence of gene B but read in another reading frame. We chose to entangle genes encoding phenotypes that would be relatively easy to test, like the ability to survive in the presence of antibiotics and the production of a fluorescent protein. Here we chose to entangle a gene conferring resistance to an aminoglycoside antibiotic with a gene encoding GFP. In order to perform entanglement of the genes, the software CAMEOS needs alignments of the gene products with many homologs. These alignements give information to the software about regions that are more or less conserved and domains of the protein that might interact although they are not in direct proximity in the primary structure. CAMEOS identifies regions where it can introduce changes in the sequence that are less likely to change the protein structure and thus its function. At the end, Cameos suggests a list of entangled genes that we call HuGenesS, since one coding sequence is longer than the second one and sort of embraces it, as if in a loving hug.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 784
- 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 784
- 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 784
Illegal BamHI site found at 378 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 784
- 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 784
- 1000COMPATIBLE WITH RFC[1000]
Knt DNA Sequence (with GFP entangled DNA sequence in capital letters):
5’ - atgattgaacaagatggattgcacgcaggttctccggccgcttgggtggagaggctattcggatatgaATGGGCAAAGGAGACGAA CTACTGCTCGGACTCGTACCAATTCTCGTTGAGCTATCAGGGCAGGTTAACGGACATCGTTTCAGCGTATCTGGCAAAGGCACTGG AGACGCTACAAGAGGAAAGCTCACGCTTAAGCTGGCTTGCACAACAGGGAGTCTACCAGTCAGCAGTCCTGGACTCGTCACAACGC TCGGGGCGGGACTGGCTTGCTTTGGGGGAAGTCCCGACCACCGACCTGCTCACAAGTTCTTTAAATCCAGCAAACCAGAAGGATAC GTACAAGAACGCACTGCGTCGGCTAAGGACTCTGGATCCTACAAAATGCGAGCTGAAGTCAAGTTCGAAGGAGACACTCTCGCGAA CAAGATCGAACTTAGAGGCATCGACAGTAAAGAAGACGGACCTATACTCGGAACACGAAGGGAATACAACTACAACTCTCACAACG CGTATATCAGCGCGGATGCCCAAAAAAACGGACTTAAAAGTAACTTCAAGAGACGCATCAACATCGAAGATAATGGTGTACAACTC GCGGACCACTACCAACAACGACTGCCTATCGGAGACGGTCCAGTACTCCTACCAGACAATCACTATCTATCCACGCAATCTCAACT CGGAGCTGACGGGAACGAGCGCAGATCGCATCTTGTACTACTACGGTTCGTCACAGCCGCAGGCATCACGCATATACTTCTACCGC TTATTGGATGAattcttctaa - 3’
Knt protein sequence resulting from the entanglement :
MIEQDGLHAGSPAAWVERLFGYEWAKETNYCSDSYQFSLSYQGRLTDIVSAYLAKALETLQEESSRLSWLAQQGVYQ SAVLDSSQRSGRDWLALGEVPTTDLLTSSLNPANQKDTYKNALRRLRTLDPTKCELKSSSKETLSRTRSNLEASTVKK TDLYSEHEGNTTTTLTTRISARMPKKTDLKVTSRDASTSKIMVYNSRTTTNNDCLSETVQYSYQTITIYPRNLNSELTGT SADRILYYYGSSQPQASRIYFYRLLDEFF
GFP protein sequence resulting from the entanglement :
MGKGDELLLGLVPILVELSGQVNGHRFSVSGKGTGDATRGKLTLKLACTTGSLPVSSPGLVTTLGAGLA CFGGSPDHRPAHKFFKSSKPEGYVQERTASAKDSGSYKMRAEVKFEGDTLANKIELRGIDSKEDGPILG TRREYNYNSHNAYISADAQKNGLKSNFKRRINIEDNGVQLADHYQQRLPIGDGPVLLPDNHYLSTQSQL GADGNERRSHLVLLRFVTAAGITHILLPLIG
Cloning our biobrick with entangled genes knt-gfp
To characterize the knt-gfp entangled genes, the DNA fragment encoding the two fully overlapping CDS in different reading frames has been inserted in the plasmid pBAD24-MoClo. The entangled genes are under the control of a promoter inducible with arabinose.
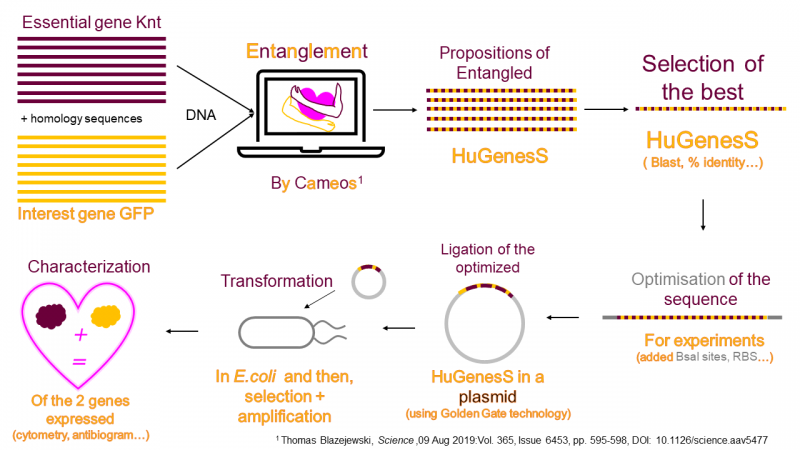
In silico assessment of the biobrick sequence with BLAST
HuGenesS sequences were analyzed to: (1) verify the presence of two ORFs, and (2) check that their products retained high identity with the original sequences. The largest CDS, that we call knt, encodes a 264 aa protein homologous to an aminoglycoside phosphotransferase (61% similarity, 52% identity). See Fig. 2.
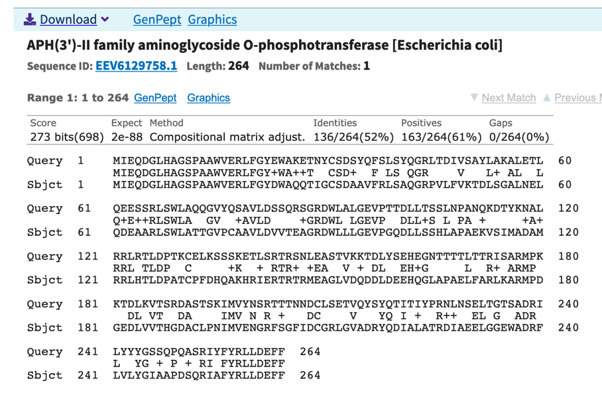
The second CDS, that we call gfp, encodes a protein with 231 residues with very high homology to a fluorescent protein (80% similarity, 74% identity) (Fig. 3).
To test whether these entangled genes encode functional proteins, they were cloned in a plasmid.
Cloning the biobrick in plasmid pBAD24-MoClo
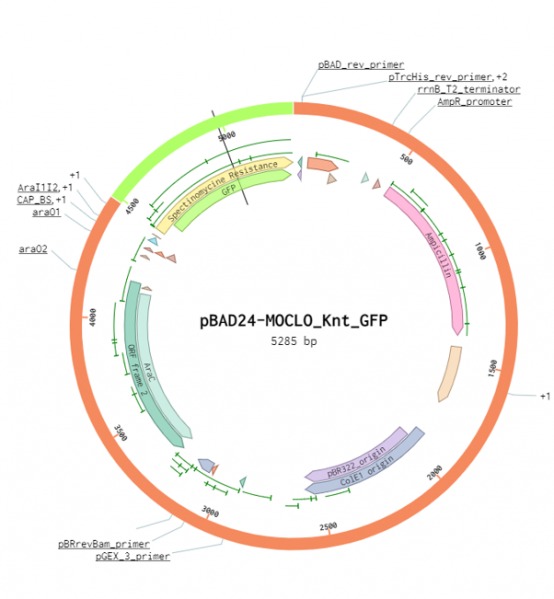
The pBAD24-MoClo cloning vector contains an ampicillin resistance gene, the pBR322_origin for replication, an araC gene encoding the AraC transcriptional regulator and the arabinose-inducible PBAD promoter (also called Para). The plasmid pBAD24-MoClo has two BsaI sites downstream of the PBAD promoter to facilitate cloning using the Golden Gate technique. It allows type IIs cloning of a coding sequence (CDS) with the prefix AATG and the suffix GCTT. The knt_gfp entangled genes were inserted in pBAD24-Moclo using the two BsaI sites to obtain the plasmid pBAD24-Knt_GFP (Fig. 1). In the absence of arabinose, AraC forms a homodimer that represses transcription of the gene beneath the control of PBAD. Arabinose modifies the conformation of AraC homodimer to activate PBAD-dependent transcription. AraC synthesis is also controlled by the presence of glucose in the media. At low glucose concentration, the adenylate cyclase produces cAMP, which associates with CRP to activate araC transcription. Therefore, maximum expression from the promoter PBAD is reached when bacteria are grown in the absence of glucose and the presence of arabinose.
Characterization Protocol
A DNA fragment encoding Knt and GFP was cloned using Golden Gate assembly into the pBAD24-MoClo plasmid. The resulting plasmid pBAD24-knt-GFP was sequenced to verify the correct sequence had been cloned, thereby ensuring that the designed plasmid was obtained. To characterize the fluorescence produced by this protein, E. coli strain DH5-alpha carrying pBAD24-knt-GFP was grown in LB supplemented with ampicillin (100 micrograms/mL) and arabinose (0.2 %). For the negative control, we used DH5alpha cells carrying the empty vector pBAD24-MoClo. As a positive control we used E. coli DH5alpha cells carrying plasmid pTH3 (de Jong et al. 2016, Scientific Reports, DOI: 10.1038/srep43889), which encodes a classical YFP (Venus). Bacteria in exponential phase were diluted in phosphate buffered saline and analyzed by flow cytometry in two separate experiments. In the first experiment we used a Partec CyFlow ML cytometer, with blue laser excitation at 488 nm and emission filter at 536 nm. In the second experiment we used a Partec cube1 cytometer with a 20 mW blue laser excitation at 488 nm and emission was measured at 536 nm. We analyzed 10000 cell counts. To assess antibiotic resistance, we either plated the bacteria on LB agar supplemented with arabinose (0.2 %), ampicillin (100 micrograms/mL) and different concentrations of aminoglycoside antibiotics, ie either kanamycin (200 micrograms/mL), spectinomycin (50 micrograms/mL) and streptomycin (50 micrograms/mL). We also tested bacterial growth on LB agar plates with antibiotic gradients. To generate the gradients, two layers of LB agar were poured in a square Petri dish. For the first layer we poured molten LB agar containing ampicillin and one of the tested aminoglycoside antibiotics in a dish that was propped up on one side by a 5° angle. Once solidified, the Petri dish was laid flat before addition of a second layer of molten agar containing only LB ampicillin. When indicated, arabinose (0.2 %) was added to both layers to ensure a constant concentration.
Result 1 : Assessing resistance to aminoglycoside antibiotics using gradient agar plates
Since the sequence Knt-GFP with the entangled genes encodes a protein with only 52% identity to an aminoglycoside modifying enzyme specific for kanamycin (Fig. 2), we performed antibiograms to evaluate whether Knt provided resistance to kanamycin but also tested spectinomycin and streptomycin in case it had changed specificity. E. coli strains carrying pBAD24 derivatives were struck on antibiotic gradient plates (Fig. 5).
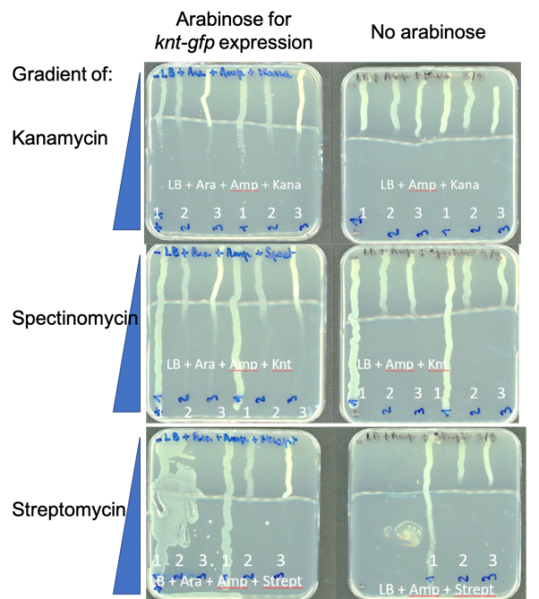
1: DH5alpha(plasmid conferring SpecR), positive control for spectinomycin resistance
2: DH5 alpha(pBAD24-MoClo-Knt-GFP)
3: DH5 alpha(pBAD24-MoClo), negative control
Growth of the positive control, a strain resistant to spectinomycin and streptomycin was observed along the length of the plates. In contrast, neither the negative control (empty vector), nor the strain carrying knt-gfp was able to grow well beyond the area of lowest antibiotic concentration, even when arabinose was included in the medium to induce gene expression.
Taken together, our results indicate that the knt gene entangled with gfp does not confer resistance to kanamycin, spectinomycin or streptomycin. It is possible that the knt gene is expressed too lowly and/or that its gene product is not functional.
Result 2 : Assessing the fluorescence of bacteria by flow cytometry
Bacteria were grown to exponential phase and analyzed by flow cytometry on two separate occasions, using different flow cytometers to widen the options of excitation wavelengths and emission filters.
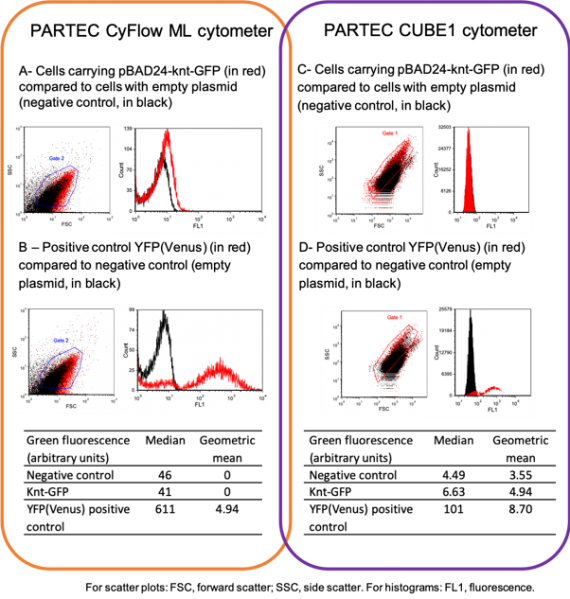
Unlike the results obtained with the positive control (panel B and D), we could not observe a significant difference in cell fluorescence for cells carrying the entangled genes knt-gfp and the negative control (red and black corresponding to Knt_GFP and negative control in panels A and B) Therefore, the GFP encoded by the entangled genes is either not functional or not produced. Additional excitation and emission wavelengths were also tested without success.
General Conclusion
We cloned a DNA fragment with entangled knt and gfp genes, but unfortunately the genes are either not expressed or their products are not functional. Expression could be further assessed by RT-qPCR and the protein might be detectable by mass spectrometry. The choice of two genes of almost the same size might have added too many constraints and could explain why the Knt is so dissimilar (52 % identity) from other proteins conferring resistance to aminoglycosides. Future work should test additional entangled genes found by Cameos.
| None |
