Part:BBa_K3427000
Heat-stable fluorescent protein YFP_LOV
The thermostable flavin-based fluorescent protein stable_YFP_LOV was originally described as a domain of a bigger protein with histidine kinase activity from the Chloroflexus aggregans, a thermophilic phototrophic bacterium (A thermostable flavin-based fluorescent protein from Chloroflexus aggregans: a framework for ultra-high resolution structural studies Nazarenko et al. 2019, Photochem Photobiol Sci, doi: 10.1039/c9pp00067d). The sequence predicted to confer fluorescence was cloned along with a short fragment encoding a tag of 6 histidines. The His-tag could be useful in the future for purification purposes. The gene was cloned in the plasmid pBAD24-MoClo under the control of an arabinose-inducible promoter. Bacteria carrying this plasmid exhibited a green fluorescence when grown in the presence of arabinose.
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
stable_YFP_LOV (113 amino acids + stop codon) :
MASGMIVTDAGADQPIVFVNRAFSTITGYAPNEVLGRN ARFLQGPQTDAATVARLREAIAAARPIQERILNYRKDG QPFWNQLSISPVRDETGNVVAFVGVQTDVTAHHHHHH*
Cloning of stable_YFP_LOV
To characterize the stable_YFP_LOV, its CDS has been inserted, with the Golden Gate technology in a plasmid under the control of an inducible promoter. This plasmid was then used to transform bacteria which will express the YFP for the study.
YFP_LOV DNA Sequence + BsaI sites :
5' - aaaaaaaaaggtctcaaatggccagtggtatgattgtgacggatgcgggtgctgacc aaccaattgtgtttgtgaaccgcgcgtttagcactataacaggctatgcacccaacg aggtcctgggacgtaacgcgagattccttcagggcccgcaaaccgacgccgctacgg ttgcccgtttacgtgaagcaatcgcggcggcacgaccgatccaggaacgcatcctga attaccgcaaagatggccagccgttttggaatcagttgtccatttcgcctgttcggg atgaaaccgggaatgtcgtagctttcgttggtgtacagaccgatgtgactgcccatc atcatcaccaccactaagcttagagaccaaaaaaaaa - 3'
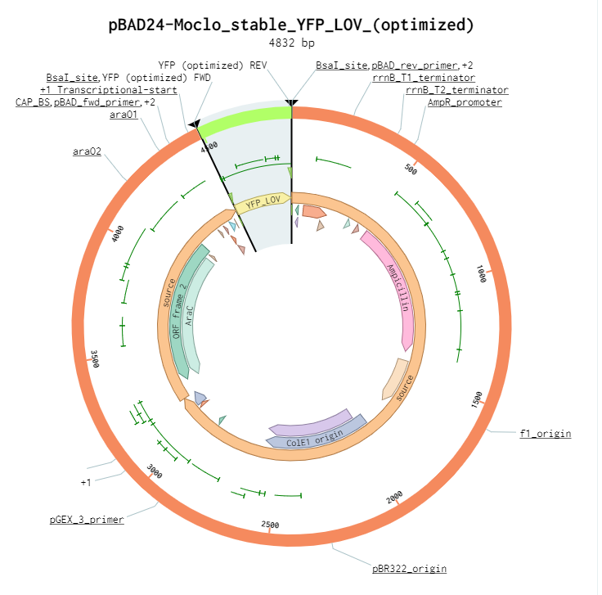
Plasmid Description
The pBAD24-MoClo cloning vector contains an ampicillin resistance gene, the pBR322_origin for replication, an araC gene encoding the AraC transcriptional regulator and the arabinose-inducible PBAD promoter (also called Para). It also has two BsaI sites downstream of the PBAD promoter to facilitate cloning using the Golden Gate technique. This feature allows type IIs cloning of a coding sequence (CDS) with the prefix AATG and the suffix GCTT. The gene encoding stable_YFP_LOV was inserted in pBAD24-Moclo using the two BsaI sites to obtain the plasmid pBAD24-YFP_LOV. In the absence of arabinose, AraC forms a homodimer that represses transcription of the gene beneath the control of PBAD. Arabinose modifies the conformation of AraC homodimer to activate PBAD-dependent transcription. AraC synthesis is also controlled by the presence of glucose in the media. At low glucose concentration, the adenylate cyclase produces cAMP, which associates with CRP to activate araC transcription. Therefore, maximum expression from the promoter PBAD is reached when bacteria are grown in the absence of glucose and the presence of arabinose.
Characterization Protocol
A DNA fragment encoding the stable_YFP_LOV was cloned using Golden Gate assembly into the pBAD24-MoClo plasmid. The resulting plasmid pBAD24-stable_YFP_LOV was sequenced to verify the correct sequence had been cloned, thereby ensuring that the designed plasmid was obtained. To characterize the fluorescence produced by this protein, E. coli strain DH5-alpha carrying pBAD24-stable_YFP_LOV was grown in LB supplemented with ampicillin (100 micrograms/mL) and arabinose (0.2 %). For the negative control, we used DH5alpha cells carrying the empty vector pBAD24-MoClo. As a positive control we used E. coli DH5alpha cells carrying plasmid pTH3 (de Jong et al. 2016, Scientific Reports, DOI: 10.1038/srep43889), which encodes a classical YFP (Venus). Bacteria in exponential phase were diluted in phosphate buffered saline and analyzed by flow cytometry in two separate experiments. In the first experiment we used a Partec CyFlow ML cytometer, with blue laser excitation at 488 nm and emission filter at 536 nm. In the second experiment we used a Partec cube1 cytometer with a 20 mW blue laser excitation at 488 nm and emission was measured at 536 nm. We analyzed 10000 cell counts.
Results
Bacteria were grown to exponential phase and analyzed by flow cytometry on two separate occasions, using different flow cytometers to widen the options of excitation wavelengths and emission filters (although eventually highest fluorescence signal was detected with excitation at 488 nm and emission at 536 nm).
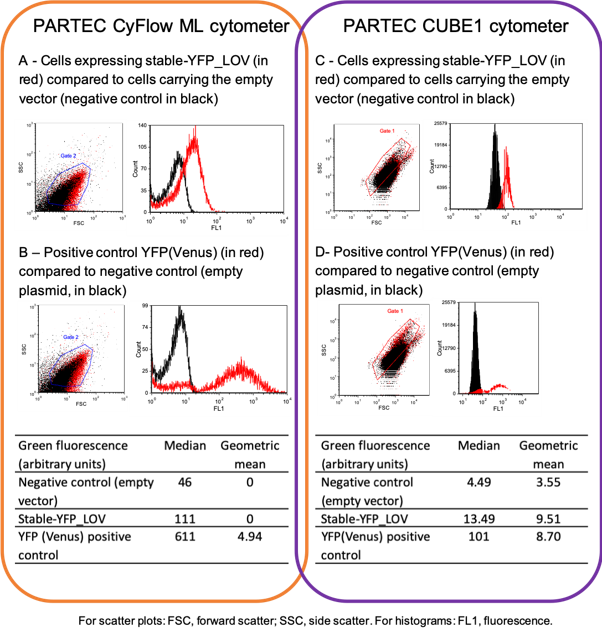
Using both cytometers we could detect fluorescence in cells producing the positive control YFP(Venus), as expected (see panel B and D). Most importantly, the fluorescence of cells producing the stable_YFP_LOV was also clearly above background (see panel A and B). Therefore, our cloned briobrick stable_YFP_LOV confers fluorescence to our chassis.
Conclusions
Sequencing confirmed that we were able to clone a new biobrick, encoding a small (113 aa residues) thermostable protein with a Cterminal histidine tag. Flow cytometry experiments show that this gene confers fluorescence (excitation 488 nm, emission 536 nm). Thus, this biobrick is correctly produced and functional.
//function/reporter/fluorescence
| None |
