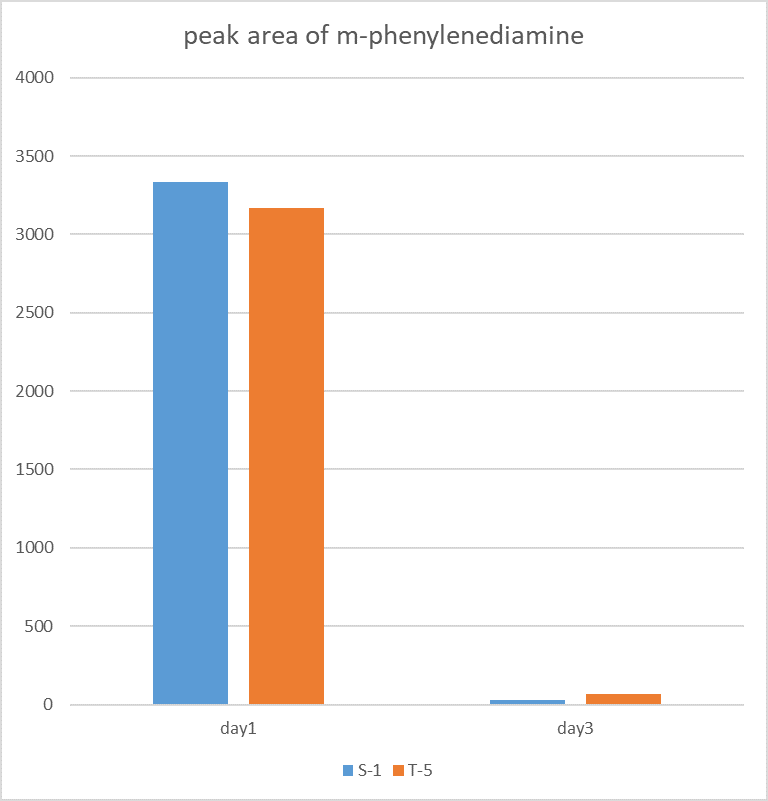Part:BBa_K3158005
J23100-RBS-bpul-ter
Bpul (Laccase from Bacillus pumilus) In the past few years, laccase has received much attention because laccases are capable of oxidizing phenolic and non-phenolic lignin-related compounds as well as highly persistent environmental pollutants. This makes them very useful for applications involving a variety of biotechnological processes. This includes detoxification of industrial wastewater, such as paper and pulp, textile and petrochemical industries. Laccase is also valuable as a medical diagnostic tool and as a tool for removing biocides from herbicides, pesticides and certain explosives in the soil. Besides, these enzymes can also be used as catalysts for the manufacture of anticancer drugs, and even as ingredients in cosmetics. Their ability to remove heterogeneous biomass and produce polymer products makes them useful tools for bioremediation purposes. We plan to express the laccase to degradate m-phenylenediamine.
Usage and Biology
BBa_K3158005 J23100-RBS-bpul-ter
(1) Transformation of Plasmid DNA
Material: Plasmid (pSB1C3-J23100-B0034-bpul-ter)
1. Take out E. coli competent cell (MG1655, DH10B, BL21(DE3)) from the refrigerator
2. Put the plasmid on ice and put it in a constant temperature water bath (42 Celsius) for 90 s
3. Use the needle tip to take a small amount of plasmid, add it to the tube with 300 μL medium, and put it onto the plate with solid media.
4. Pick the colonies in LB media and put it in shaker at 37℃
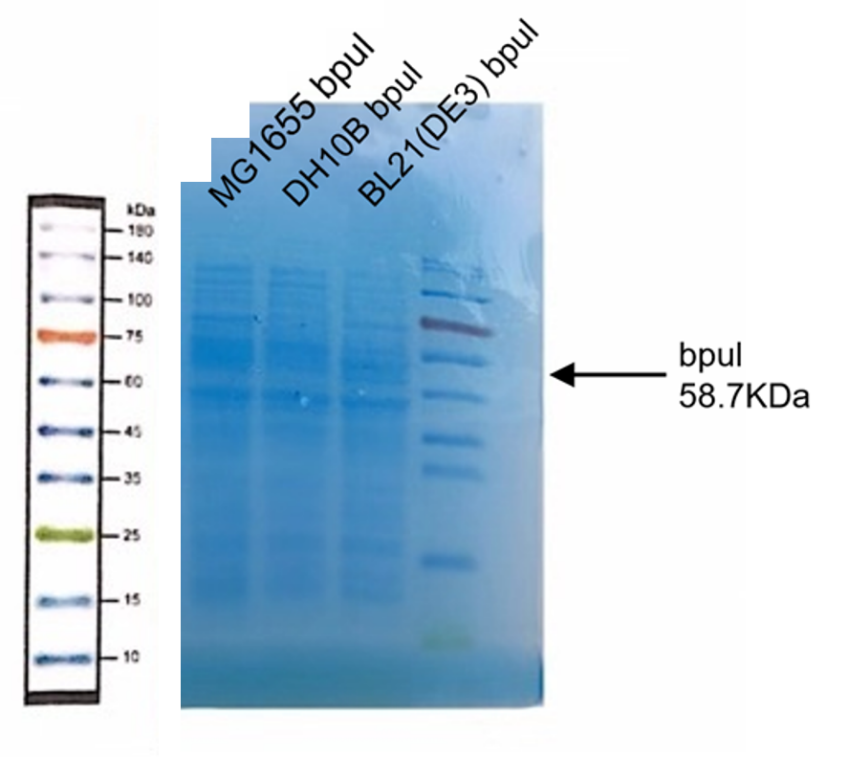
(2) Protein Electrophoresis
Material: MG1655 bpul, DH10B bpul, BL21(DE3) bpul, loading solution (6 μL), deionized water (1000 μL), marker solution, Coomassie brilliant blue dye.
1. Add 24 μL MG1655 DH10B bpul, BL21(DE3) bpul to 1.5mL tube respectively, and then add 6 μL loading.
2. Add 500 μL deionized water and centrifuge the tubes respectively.
3. Add another 500 μL deionized water, blow and suspend the solution.
4. Put the tubes 99℃ water for 15 minutes.
5. After heating, centrifuge the tubes for 5min.
6. Take and install the gel, add the electrophoresis buffer to the sample hole, and check if there is any leakage. Add marker, to the first and last sample holes, add MG1655 bpul, DH10B bpul, BL21(DE3) bpul to the other sample holes, respectively.
7. Connect the electrophoresis device to the power supply. Connect the positive electrode to the tank and the negative electrode to the slot for electrophoresis. The voltage is adjusted to 160V.
8. Turn off the power supply and disconnect the electrode until the bromophenol blue reaches the bottom of the release adhesive. Remove the glass sheet from the electrophoresis device and then remove the gel.
9. Soak the gel in Coomassie brilliant blue dye and dye it in a horizontal shaking bed for 15 min.
Observe the protein bands
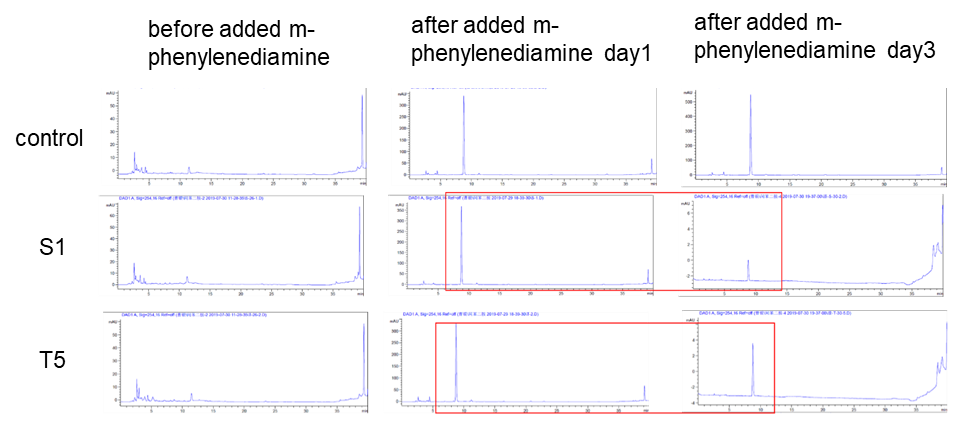
(3) HPLC
Two colonies (DH10B bpul) were picked and added with 5000 um of m-phenylenediamine, and the left side in the figure was a spectrum with nothing added. The middle was added with m-phenylenediamine for one day, and the right was after adding m-phenylenediamine for three days, the content of S-1 and T-5 was found to decrease when the content was detected.
The figure shows the peak area of m-phenylenediamine that detected for m-phenylenediamine
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 184
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 551
Illegal NgoMIV site found at 1019
Illegal AgeI site found at 307
Illegal AgeI site found at 1184 - 1000COMPATIBLE WITH RFC[1000]
| None |