Part:BBa_K3134009
Cas1-Cas2
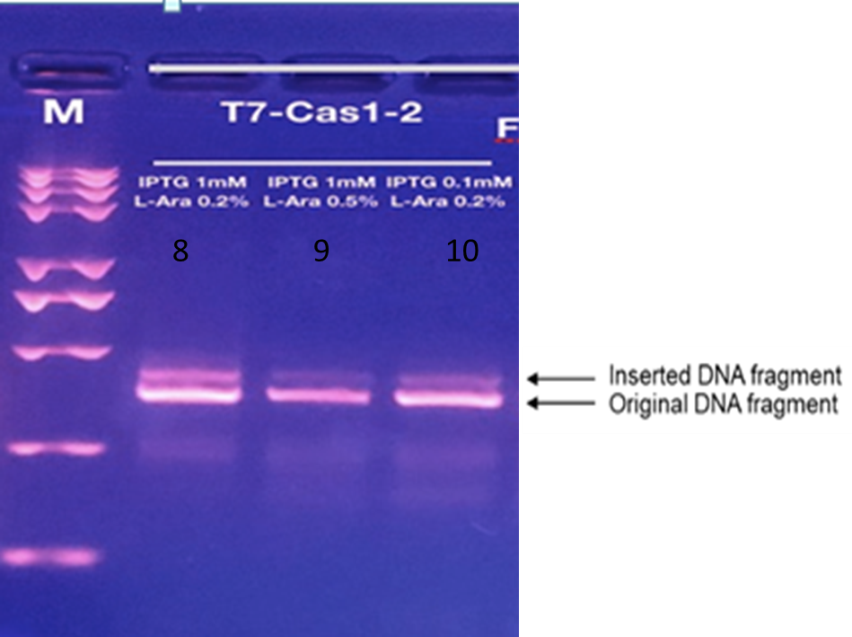
Cas1Cas2 is a protein that can insert DNA fragments into its own CRISPR array with a certain probability, which is related to the amount and time of protein expression. The analysis of agarose gel electrophoresis shows that the strip becomes longer with the presence of Cas1Cas2 because of the fragments the protein catches. With the help of this protein, we can visualize the amount of time by analyzing the brightness of the strip which appear longer than the normal one.
Usage and Biology
BBa_K3134009 Cas1-Cas2
Methods:
BL21-AI cells with pCDF-T7-Cas1Cas2:
These cells are constructed by transforming pCDF-T7-Cas1-Cas2 into BL21-AI cells. T7 is an inducible promoter that controls the gene expression downstream. Only when Isopropyl β-d-1-thiogalactopyranoside (IPTG) is added will T7 promoter starts to allow the proteins downstream, in this case Cas1 and Cas2, to express. After the expression is initiated, Cas1-Cas2 complex, the combination of E.coli Cas1 and E.coli Cas2 proteins, cuts alien DNA sequence and inserts the DNA fragments into the CRISPR array of the cells, expanding the array. Theoretically, there is a positive correlation between the acquisition of new spacers of the CRISPR array and the amount of time the expression is induced. Since the spacer acquiring ability of these cells is not influenced by blue light, it serves as the control group for the experiment. We plan to collect samples of this type of bacterial cells after the gene expression is induced for different amounts of time. PCR is then done on these samples to make more copies of the cells so that the PCR products can be run in agarose gel. We would then be able to see whether new DNA fragments are inserted into the CRISPR array under different inducing time and compare the results with those of other light detection systems.
Results:
There are many ways in which later iGEM teams can use this part. For example, in an immunity system, Cas1 and Cas2 will form a stable complex, and when a foreign DNA fragment invades the cell, Cas1-Cas2 will catch the fragment, integrate it into the CRISPR array, and form a memory, which indicates that if a team is going to introduce a immunity system against some certain diseases, Cas1-Cas2 can help a lot. Moreover, since it can catch fragments under several circumstances, a team which wants a protein to limit the expression of certain genes can use Cas1-Cas2. In conclusion, Cas1-Cas2 is of great help not only to our team, but also to later iGEM teams.
We may see that when exposed to blue light, the gel analysis will reveal a longer strip than the original one, which means we have preliminarily grasped the way to record time employees are exposed to blue light.
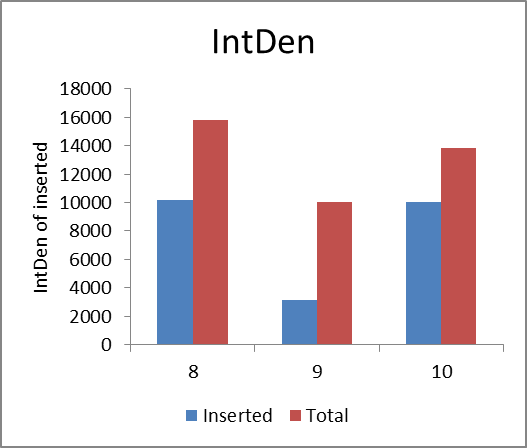
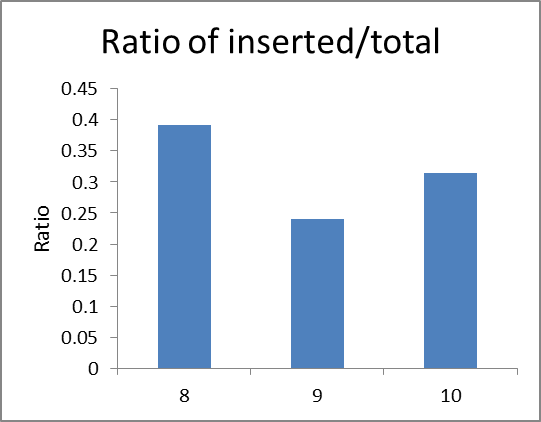
According to the light intensity of fig1, we can calculate the IntDen of inserted and total DNA fragmet and the ratio of inserted/total DNA fragment as shown in fig2 and fig3.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 116
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 837
- 1000COMPATIBLE WITH RFC[1000]
| None |