Part:BBa_K3128009
RFP protein under PLac promoter (with two restriction sites around the reporter)
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 229
Illegal BamHI site found at 913 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 789
Illegal AgeI site found at 901 - 1000COMPATIBLE WITH RFC[1000]
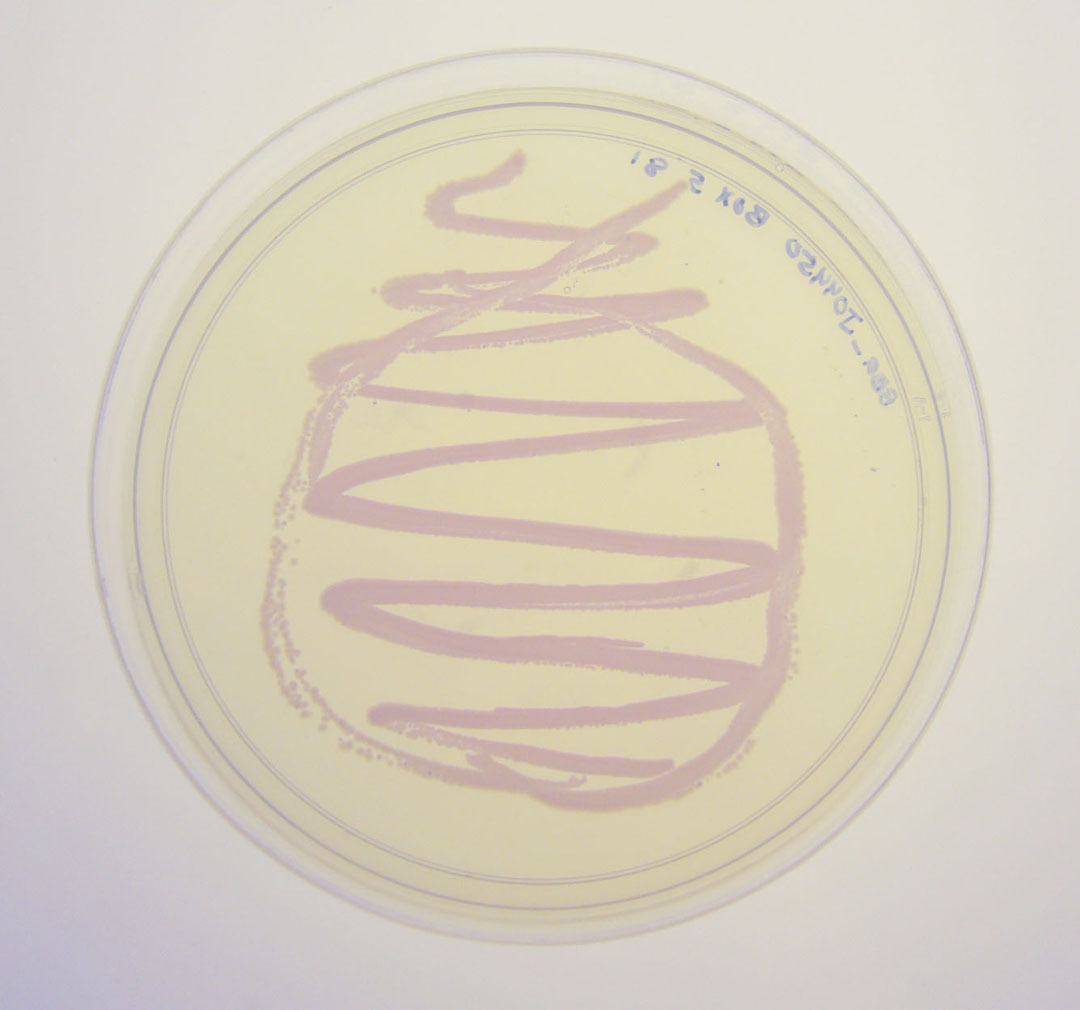
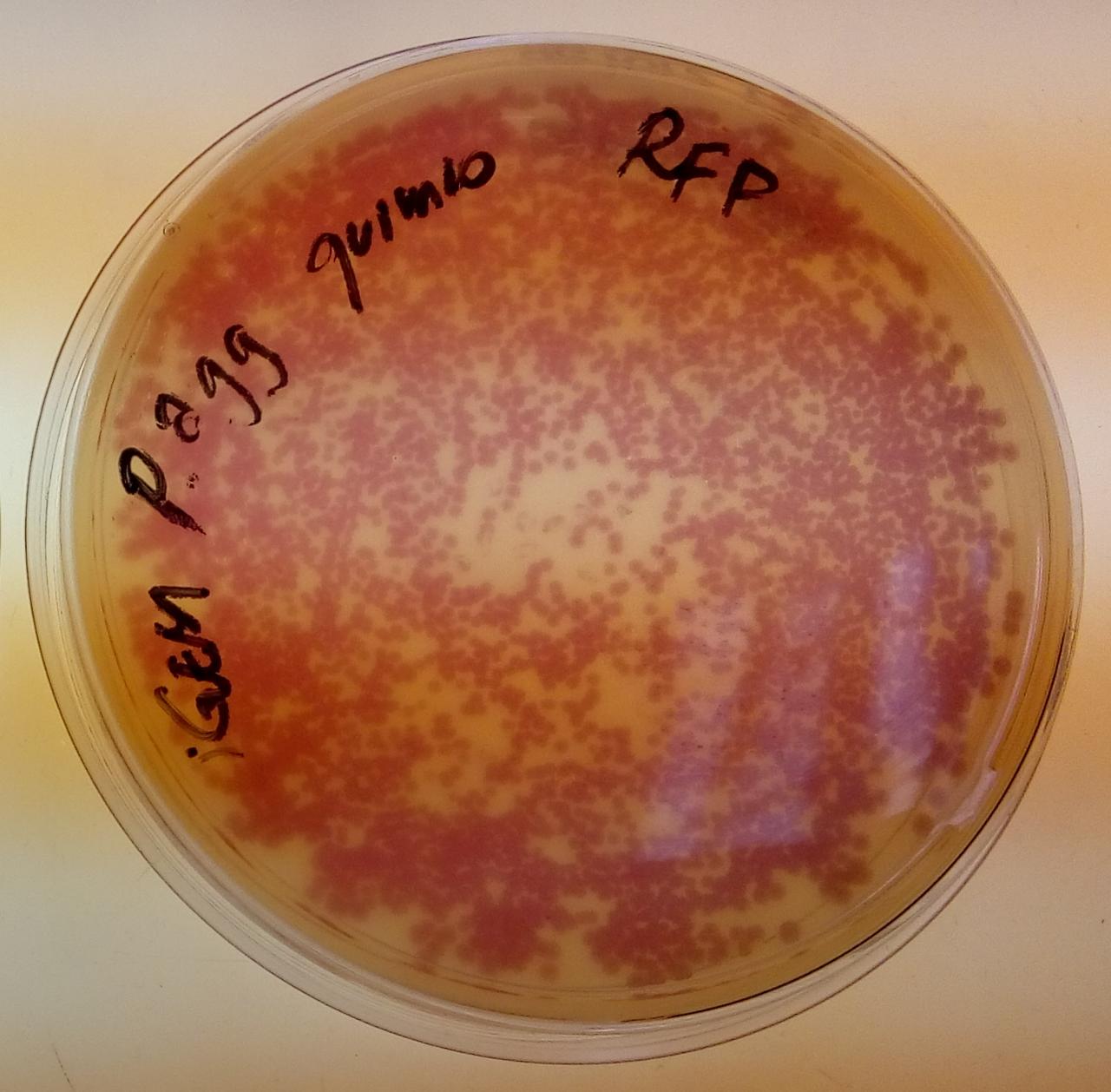
The colonies are clearly red in color under natural light after about 18 hours. Smaller colonies are visibly red under UV. The RFP part does not contain a degradation tag and the RBS is strong.
- LacI sensitive
- CAP sensitive
This part is commonly used, but can fail if the system contains LacI or CAP protein.
(--Meagan 15:39, 23 July 2009 (UTC))
[http://2012.igem.org/Team:TU_Munich Team TU_Munich 2012] improved this part by making it compatible to RFC10 and RFC25 (see: BBa_K801100)
(--VolkerMorath 15:02, 21 October 2012 (UTC))
[http://2013.igem.org/Team:NRP-UEA-Norwich Team NRP-UEA 2013] improved this part by adding a NdeI restriction site before the RFP gene. (see: BBa_K1041000)
(--holusac 20:46, 14 August 2013 (UTC))
[http://2015.igem.org/Team:Warwick Team Warwick 2015] improved this part by analysing the effect of copy number on gene expression.
(--Lcarroll 20:48, 25 September 2015 (UTC))
[http://2016.igem.org/Team:Leiden Team Leiden 2016] contributed to the characterisation of this part by showing equal functionality in simulated microgravity (0g) as in the normal gravity of the Earth.
(--Valentijn 19:38, 19 October 2016 (UTC))
[http://2017.igem.org/Team:UChicago Team UChicago 2017] contributed to this part by improving/changing the documented sequence through mutagenesis to create blunt-end restriction sites for cloning not within the prefix/suffix region (created BBa_K2428000).
(--pzulueta97 21:14, 25 October 2017 (UTC) )
[http://2017.igem.org/Team:Grenoble-Alpes Team Grenoble-Alpes 2017] contributed to the characterisation of this part by testing the time of apparition of fluorescence, in presence of IPTG or not (because the promoter leaks), as well as they contributed to the improvement of this part by using its fluorescence as a detection signal to be able to detect Vibrio Cholerae.
(--NoreenLouis 20:47, 26 October 2017 (UTC) )
[http://2017.igem.org/Team:Kingsborough_NY Team Kingsborough NY 2017] contributed to the characterization of this part by showing decreased fluorescence when expressed either in a higher salt media - such as LB with 3% sodium chloride - or E. coli that lacks tmRNA, the principal component of the cell's ribosome rescue system. View the data on the experience page or [http://2017.igem.org/Team:Kingsborough_NY/RFP visit our Wiki]
(--djcamenares 17:56, 27 October 2017 (UTC) )
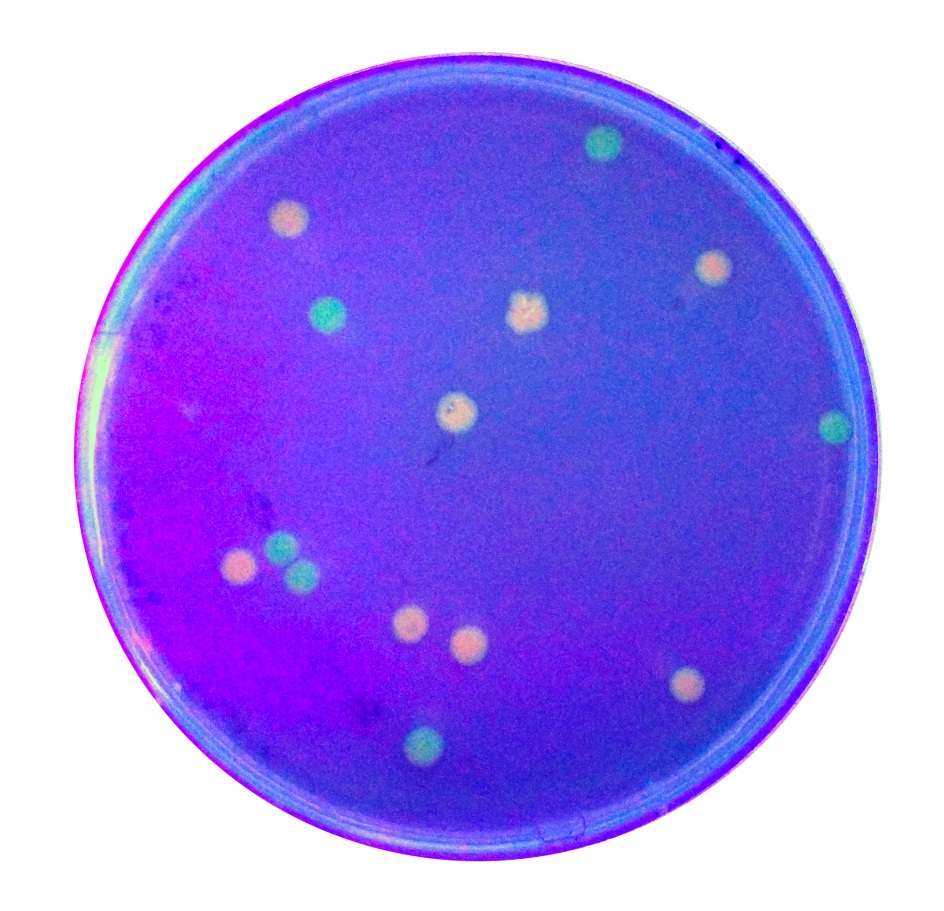
[http://2017.igem.org/Team:iTesla-SoundBio Team iTesla SoundBio 2017] contributed to the characterization of this part by analyzing the rate of false positives when using the coloring of transformed colonies as a red/white screen for determining experimental success.
(--gladish 01:26, 28 October 2017 (UTC) )
[http://2018.igem.org/Team:Grenoble-Alpes Team Grenoble-Alpes 2018] contributed to the characterisation of this part by testing the delay before apparition of fluorescence directly after transformation and the intensity of the leak, in three different E. Coli strains.
(--perrine 15:06, 9 October 2018 (UTC) )
[http://2018.igem.org/Team:H14Z1_Hangzhou Team H14Z1_Hangzhou 2018] contributed to the characterisation of this part by testing the fluorescence idensity in different condition(e.g.temperature,medium volume, IPTG concentration) ,in two different E. Coli strains.
(--ericxu 09:32, 17 October 2018 (UTC) )
[http://2018.igem.org/Team:SKLMT-China Team SKLMT-China 2018] improved this part by replacing the induced promoter lacI with the strong constitutive pf-5 promoter PampC.(see: BBa_K2569029)
(--DDY 16:47, 17 October 2018 (UTC) )
[http://2019.igem.org/Team:Grenoble-Alpes Team Grenoble-Alpes 2019] contributed to the characterisation of this part by testing the effect of cAMP on the promoter’s leakages and protein expression in Escherichia coli.
(--pinerol 19:50, 27 September 2019 (UTC) )
Pictures
Team ITB_Indonesia 2017: Red color dynamics of cloned Escherichia coli strains in LB broth

In Team ITB_Indonesia 2017
characterization, we found in normal growth/incubation condition (37 oC, LB agar) that
BBa_J04450-transformed Escherichia coli BL21 colony appear to need longer incubation time (>18
hours) until it clearly shows red color under natural light.
We then investigate whether this phenomenon is influenced by the strain, and we try if there are
lac repressor in the system that can be released by inducing the culture with IPTG, hence
increasing the expression of mRFP.
Experimental Design
We used three different strains of transformed E. coli (BL21, DH5alpha, and Top10) for this
study. They were incubated in LB broth, 37 oC, and sampled every 4 hours for 2 days to
determine the red color absorbance at 588 nm. The amount of IPTG added for respective treatment is 500
µM.
Result and Findings
- There are no significant differences of mRFP expression in different strains of E. coli (BL21, DH5alpha, Top10)
- There are no significant effects of mRFP increased expression after IPTG induction.
- The red color absorbance under 588 nm wavelength is recorded around 2.5-3 OD units.
- The broth become red in color under natural light around 16-20 hours of incubation time.


 ==Pictures==
==Pictures==


Team INSA-UPS France 2017 : usage in Vibrio harveyi strain engineered by conjugation
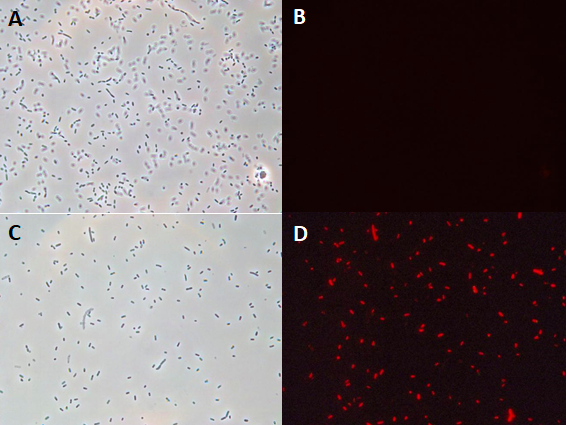
In the context of the iGEM INSA-UPS France project 2017, BBa_J04450 was tested in the Vibrio harveyi background. To the best of our knowledge, RFP has never been used in this strain. BBa_J04450 biobrick was cloned in a broad host range plasmid (pBBR1MCS-4) and conjugated into V. harveyi. The protocol of triparental mating can be found here. Its expression has been studied by fluoresence microscopy in Vibrio harveyi .
The microscopy results demonstrated the fonctional production of RFP in Vibrio harveyi, and hence, the functionality of part BBa_J04450 in this background.
IIT Madras 2016's Characterization
Experimentation
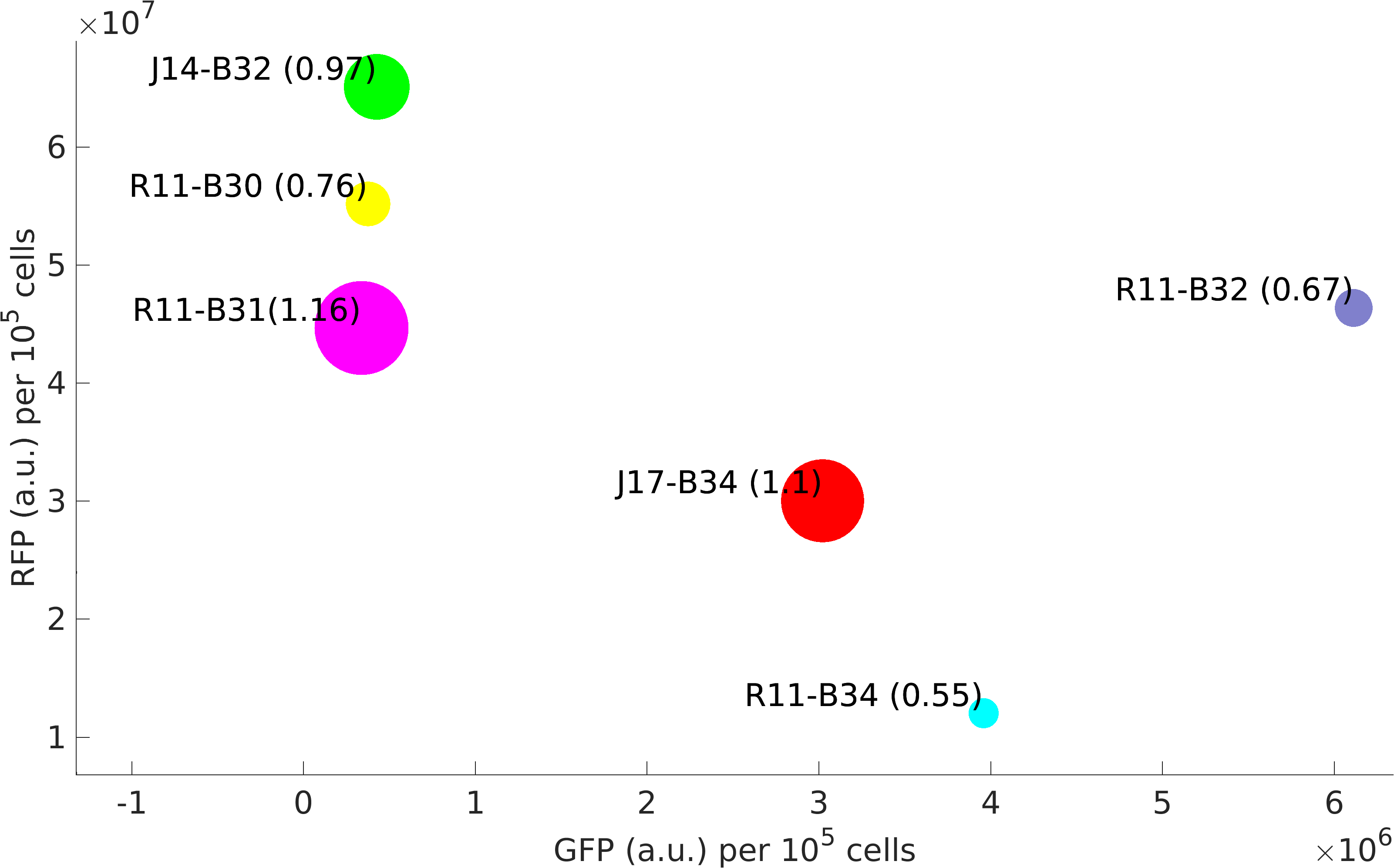
This BioBrick was used along with various GFP producing devices to understand the role of RBS and Promoter parts in giving rise to intrinsic noise in E. coli DH5alpha. Expression data for GFP and RFP proteins were obtained using flow cytometry (BD FACS Aria III) at 3hr, 6hr, 9hr and 12hr stage of growth along with cells expressing only GFP, only RFP and none. Cumulative intrinsic and extrinsic noise were measured using modified [http://2016.igem.org/Team:IIT-Madras/Model#Noise_in_Devices| Elowitz formula]. OD600 values for specific growth rate estimation were obtained using Spectrophotometer over an interval of an hour for 12 hours. Given specific growth rates are in it's logarithmic values. This BioBrick can be used to characterize noise and strength of complex devices by cloning this device with given device, which produces a different reporter protein. In graphs, we have R11-B32, R11-B34, J14-B3, J17-B34, R11-B30 and R11-B31 in pSB1A2 plasmid backbone.
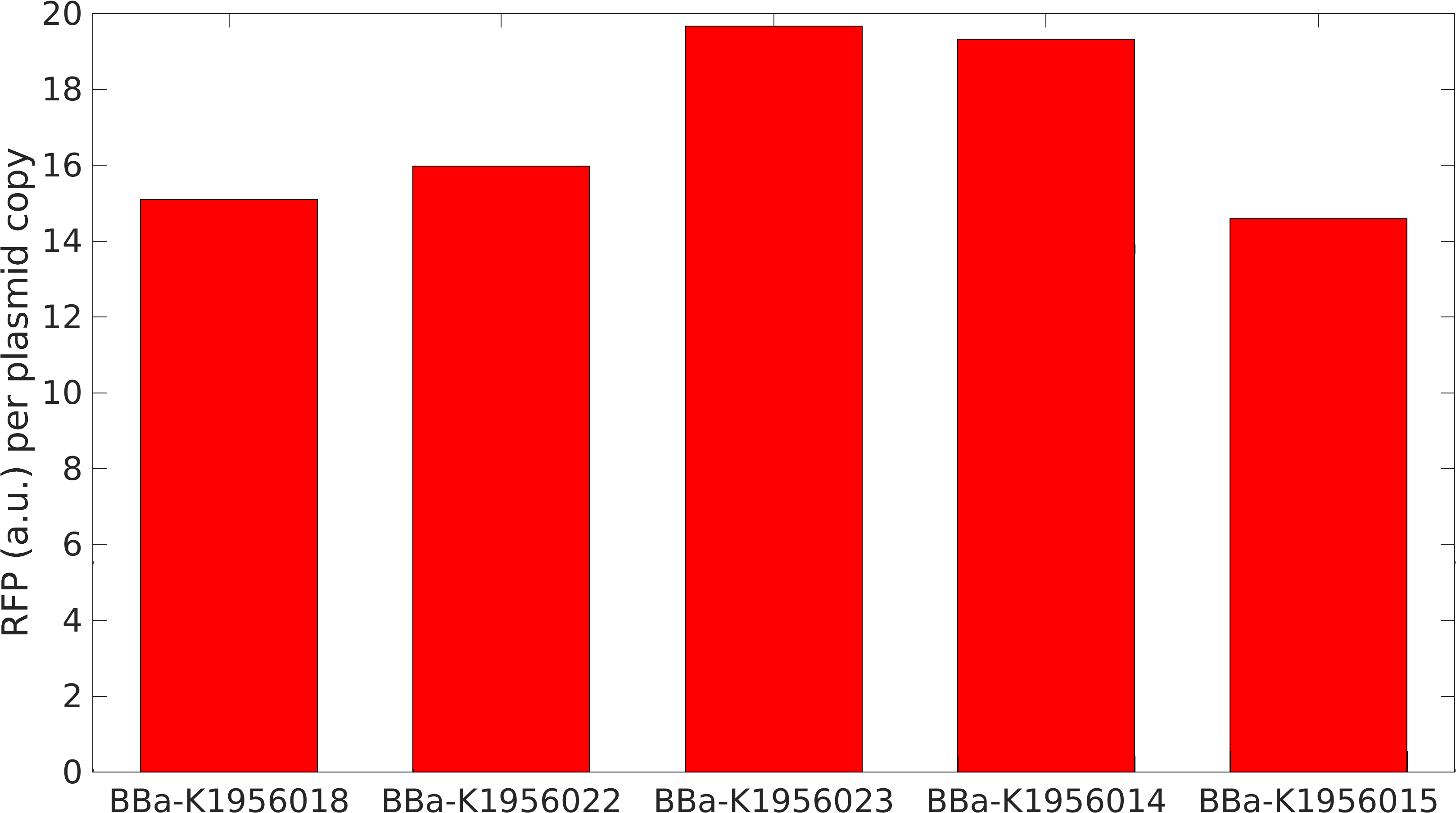
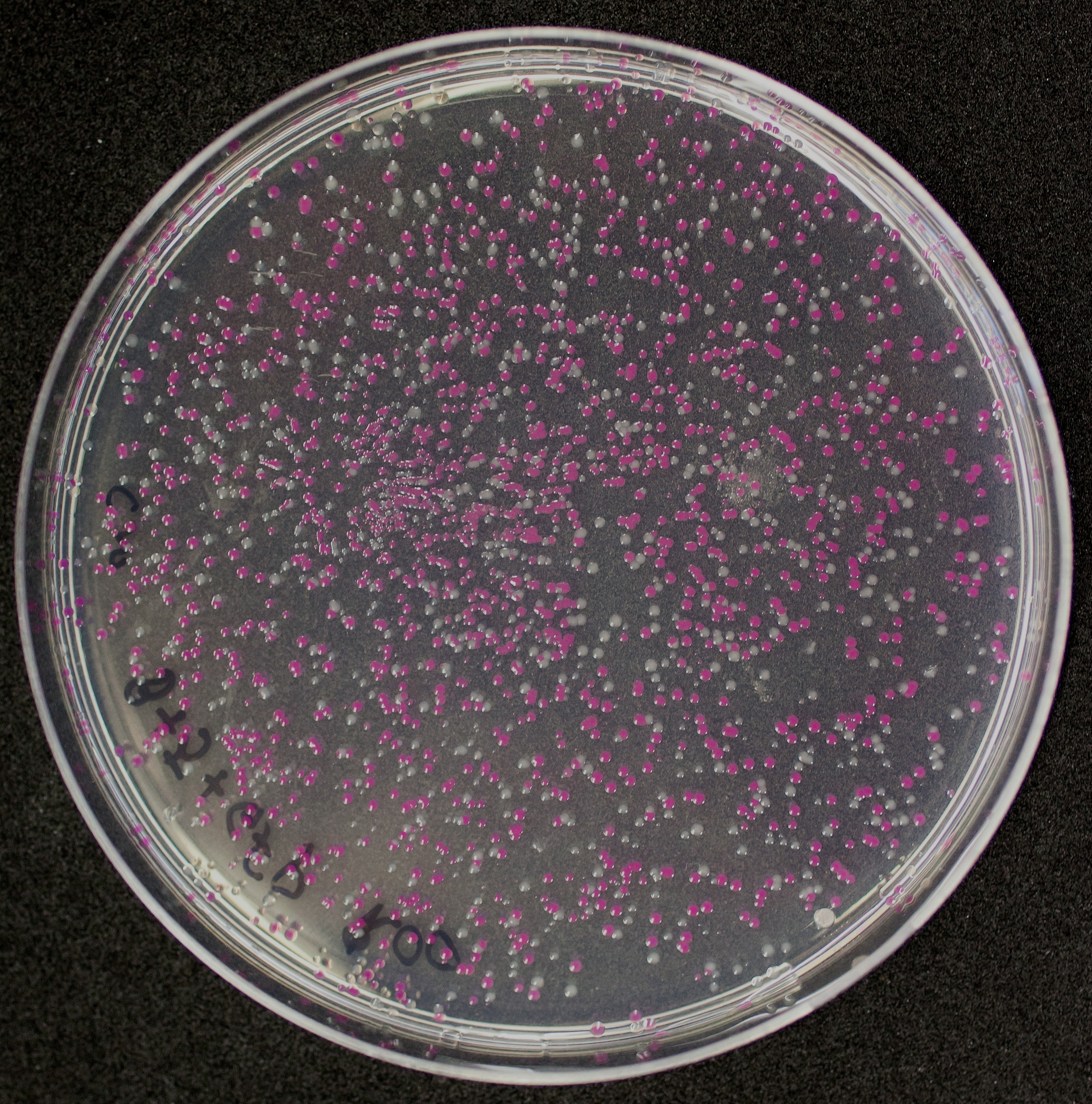
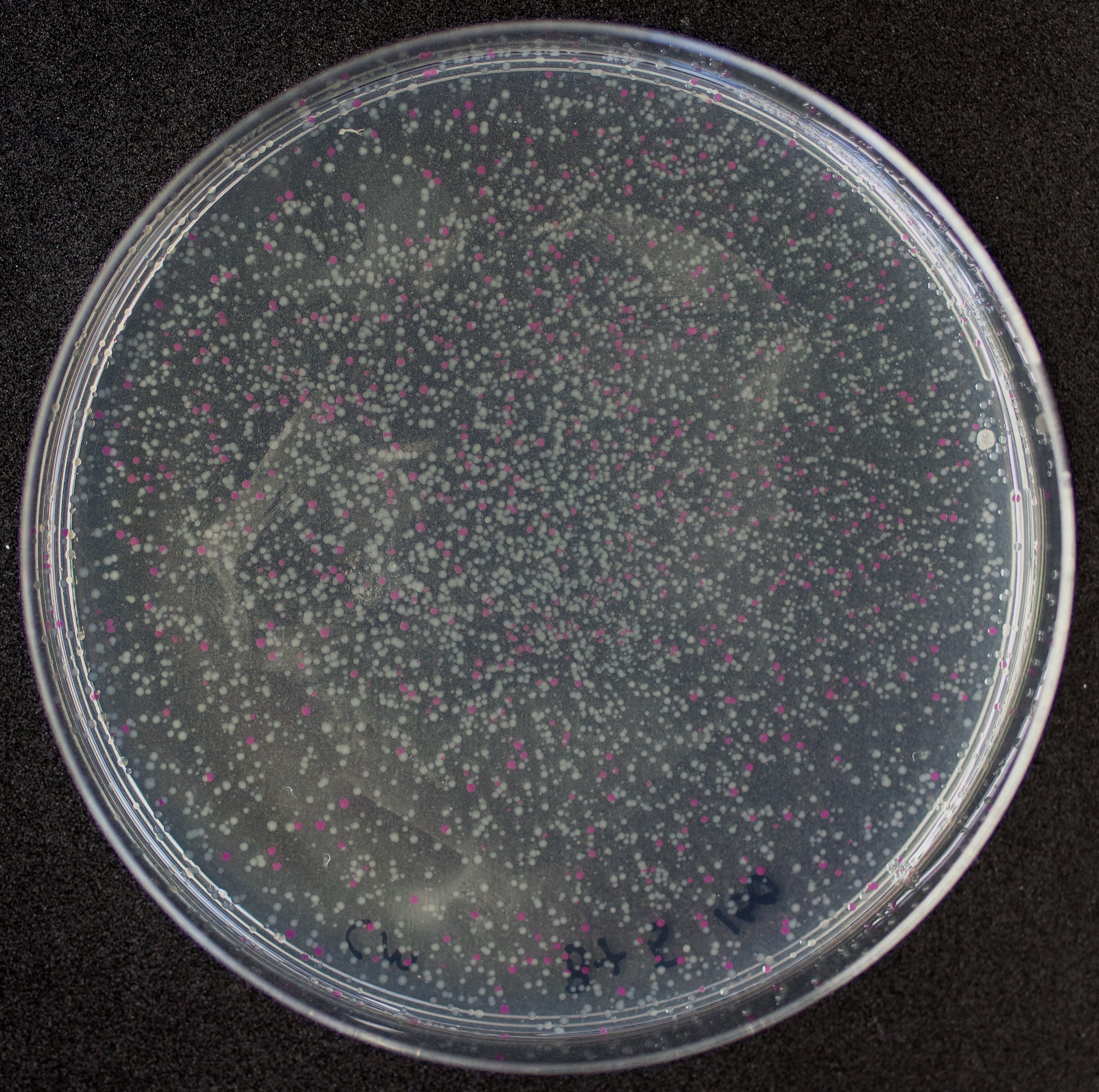
[http://2010.igem.org/Team:Groningen Team Groningen 2010] reports the usage of this part as a cloning tool. When ligating any part, or part assembly, into any standard backbone that contains this part, the non-restricted and single-restricted backbones that self-circularize will produce red colonies on rich media plates (we use TY). These undesired transformants can than be avoided in the screening for the correct construct. With this method, the backbone desired for a new construct does not need to be purified from agarose gel to decrease the amount of undesired tranformants caused by ligation of the original part present in the backbone. The amount of incorrect transformants depends, of course, on the ratio of backbone (mixed with J04450) vs. BioBrick insert, the size of the BioBrick insert, and whether the insert is an assembly of two BioBricks. The images below show two ligations with different efficiencies.
Usage in Chromobacterium Violaceum
[http://2016.igem.org/Team:Tec-Monterrey Team Tec-Monterrey 2016] characterized the output of the part BBa_J04450 in a novel chassis, Chromobacterium Violaceum, as it produces a native purple pigment Violacein, we were curious whether RFP would be useful as a reporter gene. Furthermore, we characterized its expression under lac promoter. We did the transformation of C. Violaceum by a method that has not been reported yet, we made C. Violaceum competent cells with the protocol that is in our wiki, we concluded that the best O.D. for the heat shock transformation is 0.5 since it showed clearly better results than 0.4 or 0.6, we will continue to work in the transformation efficiency.
[Image: ]
]
H14Z1_Hangzhous 2018's Characterization
[http://2018.igem.org/Team:H14Z1_Hangzhou Team H14Z1_Hangzhou 2018]
Purpose
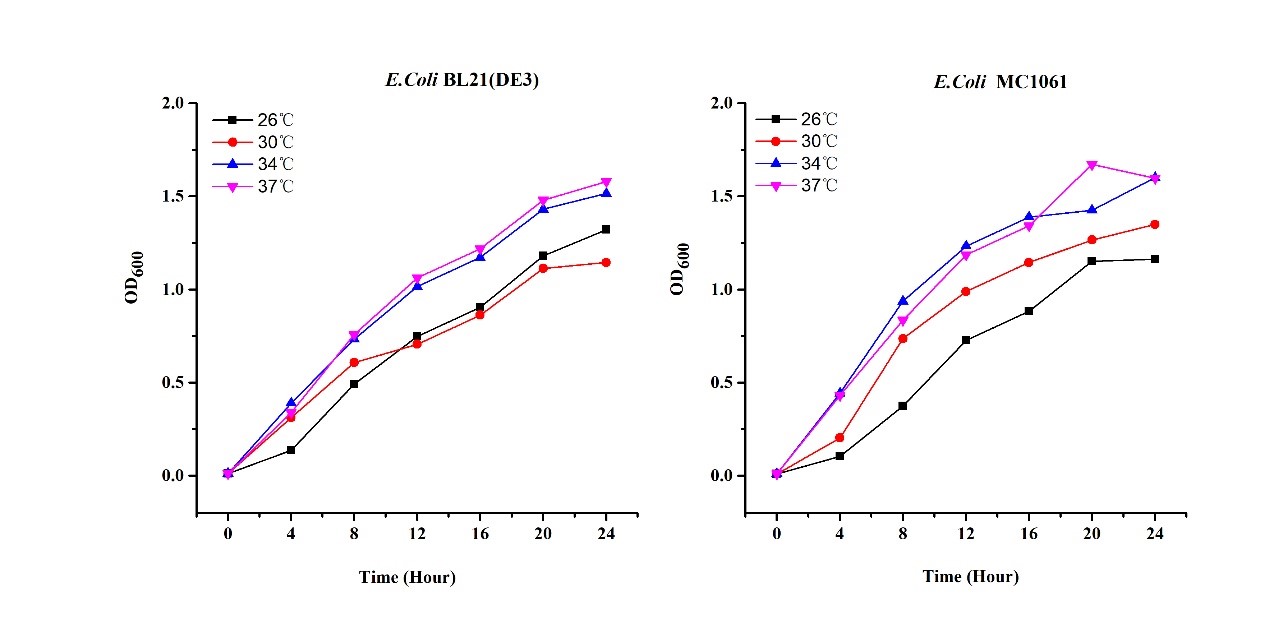
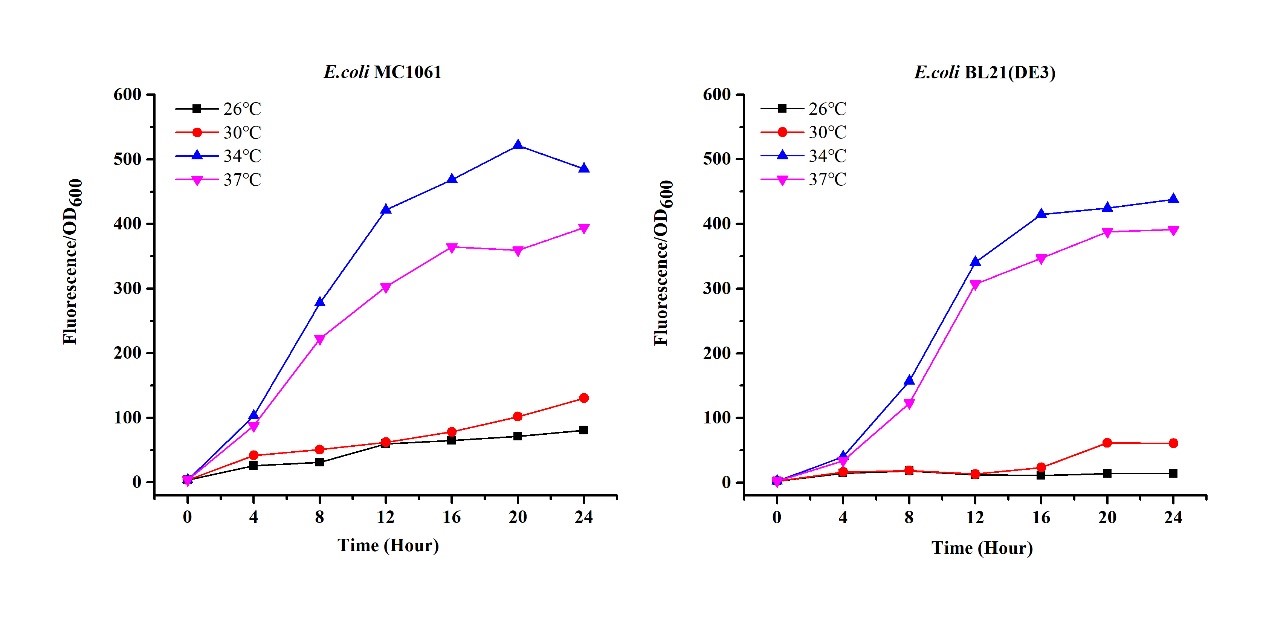
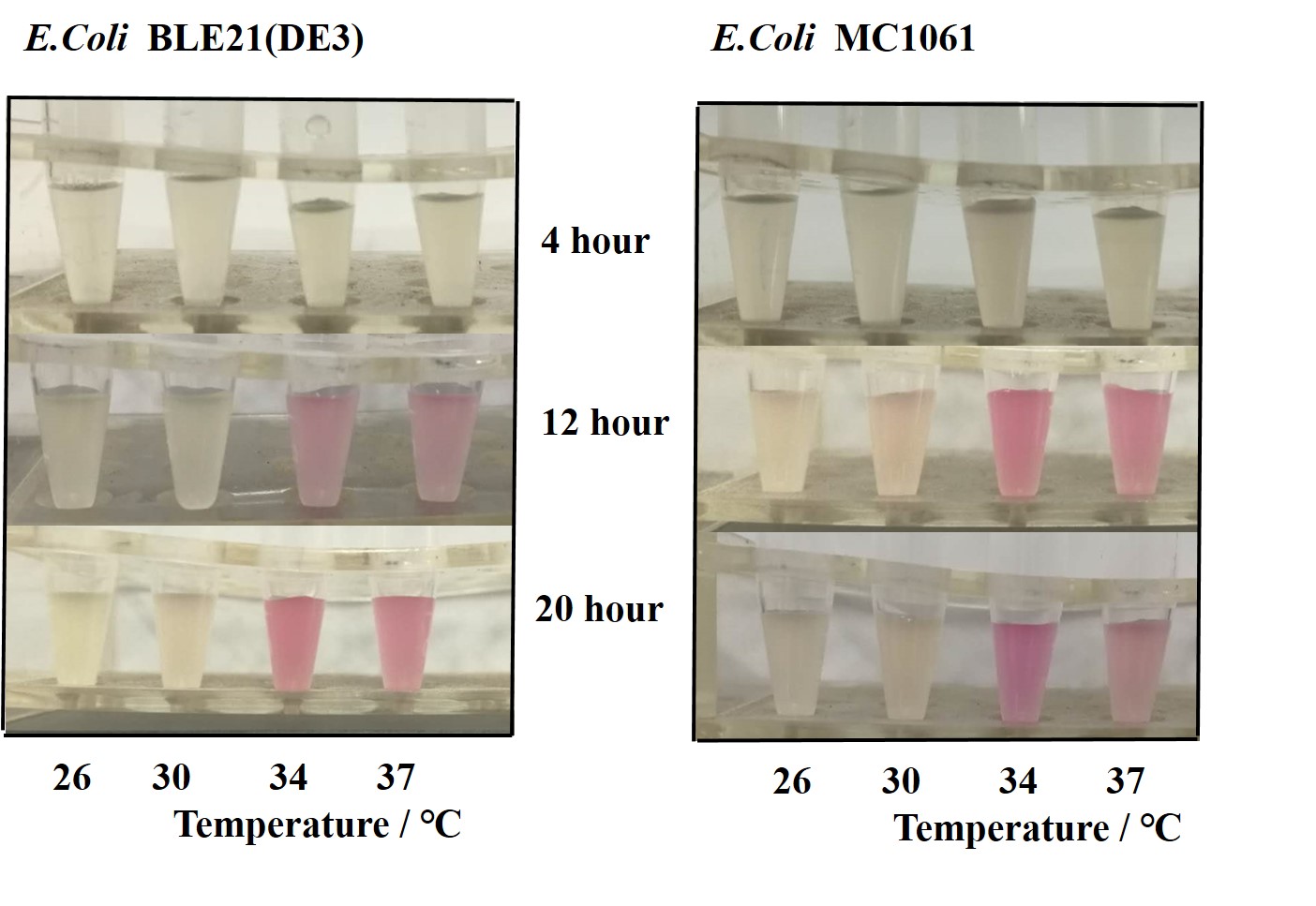
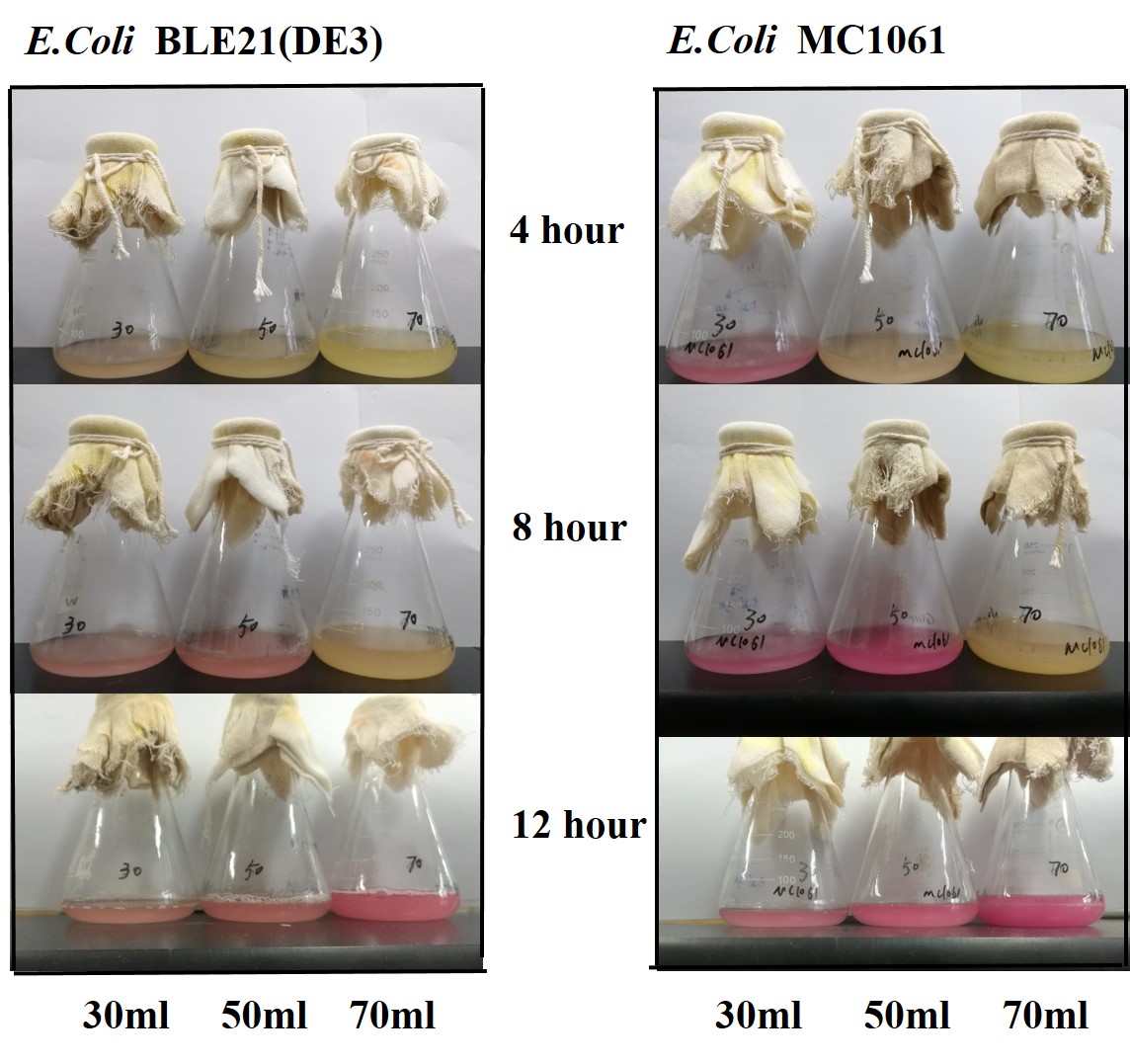
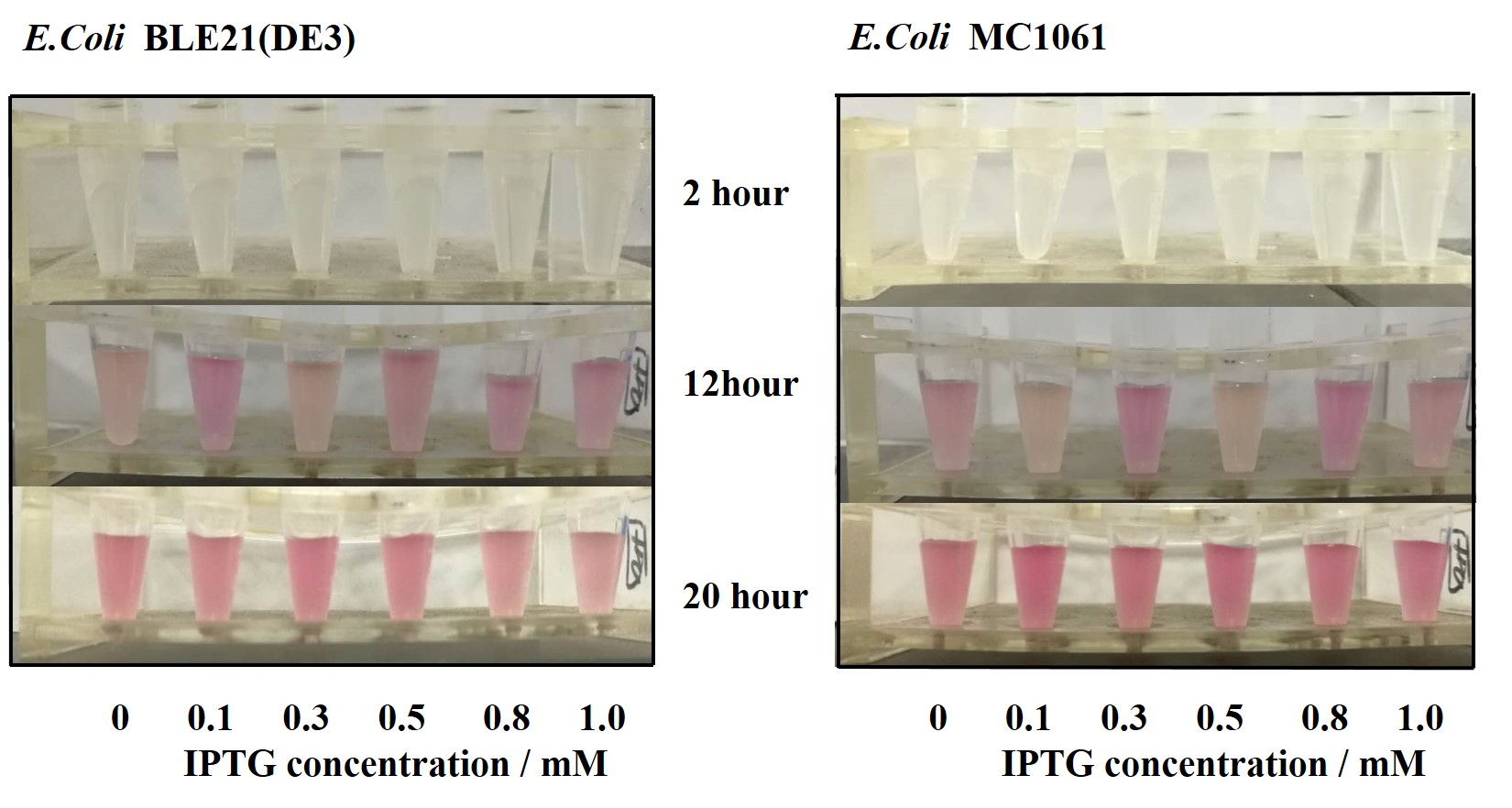
Previous experiments had shown that BBa_J04450-transformed E. coli BL21(DE3) would turn red color under natural light after 18 hours due to leaking expression. This part is useful for characterizing the expression of different proteins using the lacI sensitive promoter. For better application of this promoter in controlling protein expression, the effects of different hosts under various conditions were investigated systematically. The target hosts include E. coli BL21(DE3) and E. coli MC1061. The target culture conditions were temperature, inducer concentration and medium volume in the flask.
Experiment design
- Exploring the effect of temperatures on mRFP expression
1.Inoculate 5 ml LB with monocolony (BL21(DE3)/MC1061) from the plate. Grow the cells overnight at oC,200 rpm.
2.Dilute pre-culture 1:100 into 5ml LB (BL21(DE3)/MC1061) and cultivate at different temperature (26, 30, 34, 37 oC), 200 rpm, respectively.
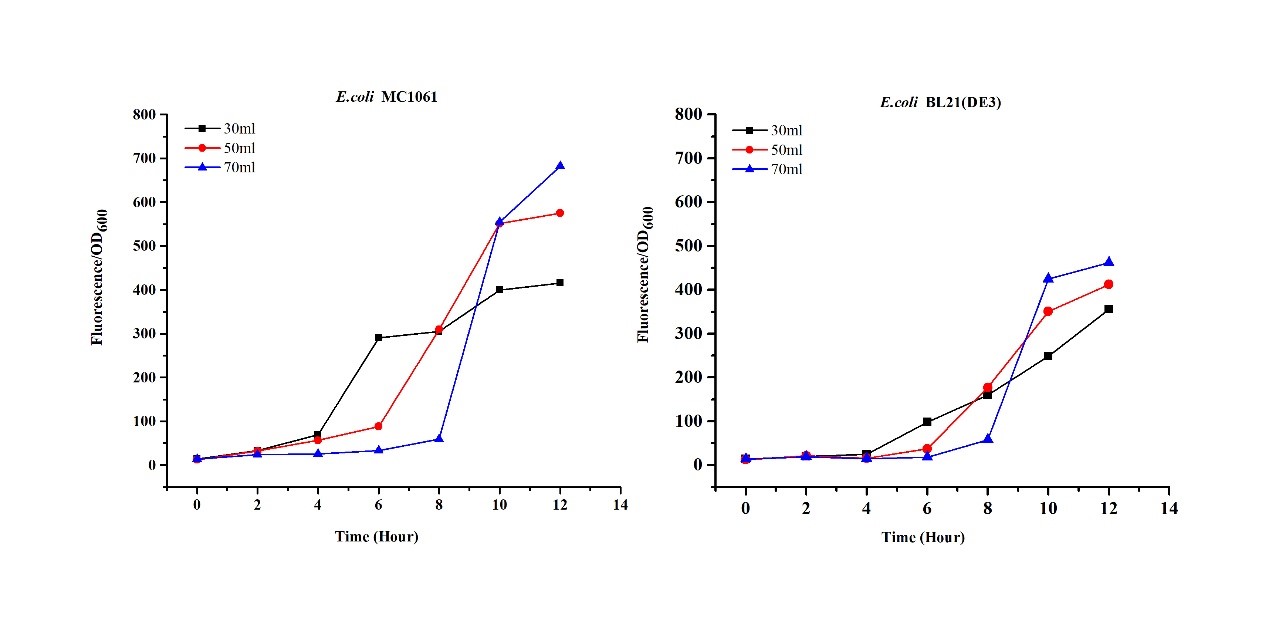
3.Measure the fluorescence and OD600 every 4 hours. - Exploring the effect of medium volume on mRFP expression
1.Inoculate 5 ml LB with monocolony (BL21/MC1061) from the plate.Grow the cells overnight at 37oC, 200 rpm.
2.Dilute pre-culture 1:100 into 30ml, 50ml, 70ml LB (BL21(DE3)/MC1061) and cultivate at 37oC, 200 rpm.
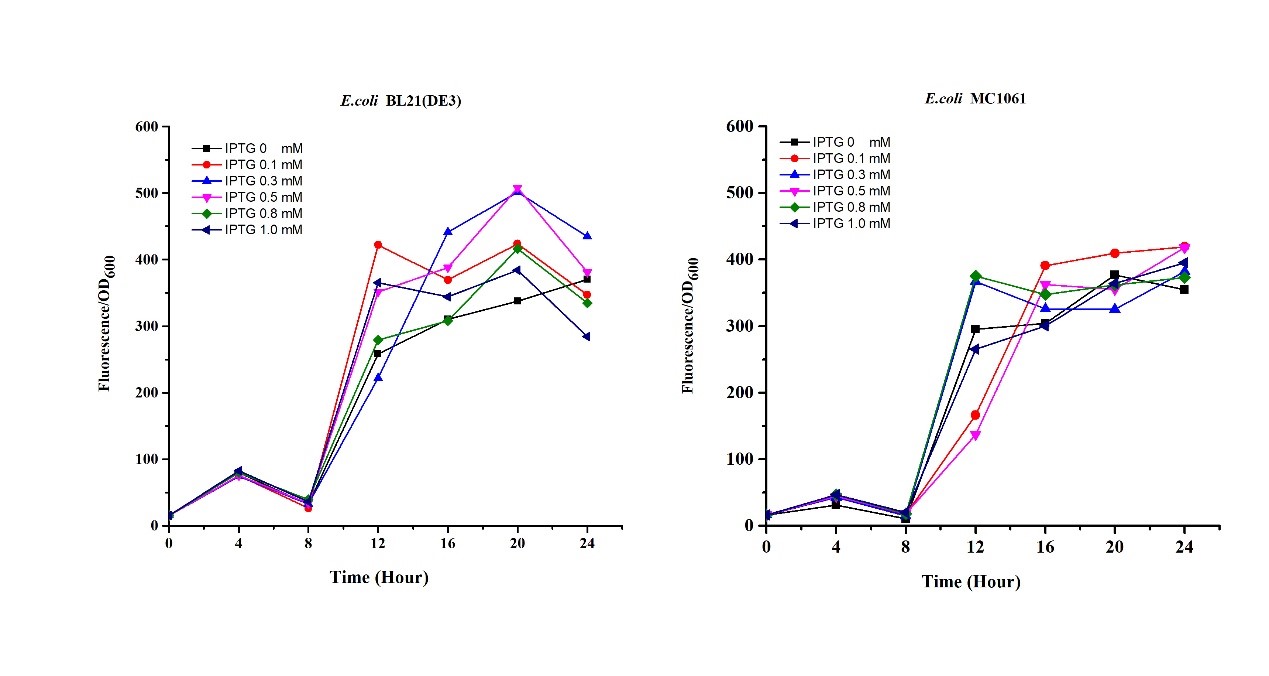
3.Measure the fluorescence and OD600 every 2 hours. - Exploring the effect of concentration of IPTG on mRFP expression
1.Inoculate 5 ml LB with monocolony(BL21(DE3)/MC1061) from the plate. Grow the cells overnight at 37oC, 200 rpm.
2.Dilute pre-culture 1:100 into 5ml LB (BL21(DE3)/MC1061), cultivate at 37oC, 200 rpm. Protein expression are induced using 0.1, 0.3, 0.5, 0.8, 1.0 mM IPTG, respectively.
3.Measure the fluorescence and OD600 every 4 hours.
Results
- Exploring the effect of temperatures on mRFP expression
- Exploring the effect of effect of medium volume on mRFP expression
- Exploring the effect of concentration of IPTG on mRFP expression
There are some differences of growth rate at different temperatures for both E. coli (BL21(DE3), MC1061) (Fig.1). However, the differences of the fluorescence intensity at different temperature is much higher than those of the growth rate at various temperatures by using two E. coli cells (Fig 2). The fluorescence intensity followed same trend in two E. coli (BL21(DE3)/MC1061).The results indicated that the leaking expression was lower in low temperature (e.g.26 and 30oC) and higher in high temperature. Highest leak appeared in 34 oC. This finding is different from our expectation, where we expected that the highest leaking expression would be at 37 oC
The mRFP expression of two strains showed the same trend in different volume of medium in the flask. After 6 hours, the fluorescence intensity in 30 ml medium-containing flask is significantly higher than others, indicating that mRFP express fastest in this volume scale (Fig 3). This suggest that dissolved oxygen is an important factor which will affect the protein expressin and cell growth, since the lower medium volume in the same type flask will bring about higher level of dissolved oxygen in the medium.
Our results showed that significant fluorescence differences were not observed by using different IPTG induction (Fig 3). However, it is interesting that the fluorescence intensity of E. coli BL21(DE3) after IPTG induction are higher than those of E. coli MC1061, not similar to the trend in our previous experiment (Fig 2). It might be explained that the expression of mRFP can be accelerated by IPTG inductin in BL21(DE3) with regard to other E. coli hosts.
Pictures
SCAU-China 2018's improvement
The BBa_K2559005 is a amended eGFP coding part improved from BBa_l714891.
Usage and Biology
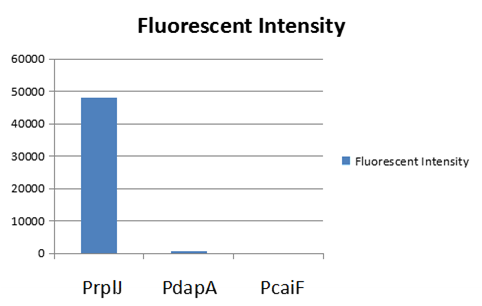
The part BBa_K2559005 has a sequence improvement on the basic part submitted by iGEM07_Peking (BBa_l714891) which encodes the SDY_eGFP. However, we found out a 16 bp nucleotides redundancy in the eGFP starting coding region in BBa_I714891, after checking the sequence of BBa_I714891 from NCBI. Therefore, we decided to delete the redundant 16 bp nucleotides in BBa_I714891 to amend the length of eGFP coding sequence. The amended eGFP coding biobrick is the BBa_K2559005. To test the function of BBa_K2559005, we designed a new E.coli expression vector containing our new part termed as BBa_K2559003under a strong E.coli endogenous promoter (PrplJ). Therefore, the amended eGFP in BBa_K2559005 was driven by PrplJ promoter, and expressed in DH10B. In addition, we also applied the BBa_K2559005 in the promoter intensity analysis of our other two new parts, the BBa_K2559004 and BBa_K2559011 which are relatively weaker E.coli endogenous promoters (PdapA and PcaiF) (Figure 1).
We summarized that our improvedpart, the amended eGFP coding biobrick BBa_K2559005 worked well in DH10B. We also hoped that our improvement on the BBa_I714891 can help their future applications by other groups in the future. However, it is difficult for us to perform additional experiments with BBa_K2559005 and BBa_I714891 due to the unavailable BBa_I714891.
To expand the application of BBa_K2559005, we searched theBBa_J04450 stored in registry and do another improvement in the BBa_J04450. The BBa_J04450 is a strong RFP expression vector in E.coli. As the main page of BBa_J04450 mentioned, the E.coli colonies with BBa_J04450 were in red color under normal light after about 18 hour culture on LB plate (Figure 2). We used the BBa_K2559005 to replace the RFP region in BBa_J04450, the modified part is BBa_K2559009. We transferred the BBa_K2559009 to DH5α by heat-shock, and found that the fluorescence signal can be observed under the UV (Figure 2).
So, we confirm that our improved part BBa_K2559005 can work in different E.coli expression system. We are also looking forward to more application of the BBa_K2559009! </html>
Grenoble-Alpes 2019 Characterization
The goal here is to characterize the effect of cAMP on the promoter’s leakages and overall proteins expression
MAIN RESULTS
DH5α which is a standard strain with endogenous adenylate cyclase
activity.
BTH101 which is a strain without endogenous adenylate cyclase activity
and so without production of cAMP. BTH101 strain is streptomycin
resistant.
BTH101 + adenylate cyclase (BTH101-Zip) to generate cAMP in the
strain.
To restore the adenylate cyclase activity we used two plasmids: one
containing pUT18-LeucineZipper (pUT18-LZ) and the other containing
pKT25-LeucineZipper (pKT25-LZ). The Leucine Zipper’s (LZ) will
homodimerize and will bring T18 and T25 closer which will restore the
adenylate cyclase activity thus allowing the production of cAMP by
BTH101. Both T18 and T25 are under the control of an IPTG inducible
promoter (lactose promoter) to create an auto amplifier system.
BBa_J04450 was cloned in pKT25-LeucineZipper, and the bacteria was co-
transformed with both plasmid T25-LeucineZipper + BBa_J04450 and
T18-LeucineZipper.

It is apparent that without IPTG there is a visible difference between
strains that can produce cAMP (DH5α and BTH101-Zip) when compared with the BTH101 strain that do not produce any cAMP.
The DH5α red colonies reveal a substantial RFP expression as the result of the
leakage of the promoter.
BTH101 has white colonies indicating that no visible RFP was produced. This data
suggest that the promoter does not leak in that condition.
BTH101-Zip presents same red colonies than the DH5α strain.

RFP expression by BTH101 with or without cAMP shows that without cAMP, the lactose promoter has almost no leakage suggesting that the leakage of the lactose promoter is due to cAMP in absence of IPTG. When the cAMP production is restored the promoter is activated and the protein is expressed.
It is also interesting to note that with the BT101-Zip there are very little protein expression if no IPTG is added.
Usage and Biology
Using cAMP depleted strains and T18-leucineZipper/T25-leucineZipper under the control of a lactose promoter is a good alternative to produce inducible proteins without leakage//classic/reporter/pret
//function/reporter/color
//function/reporter/pigment
| emission | RFP |
| excitation | |
| tag | None |