Part:BBa_K2985009
Pul-> Amylopectin 6-glucan hydrolase
Pul gene encodes a protein, pullulanase, which can specifically hydrolyze the alpha-1,6 glycoside bonds in amylopectin branching points by endonucleation, thus cutting down the whole branching structure to form amylose amylopectin debranching enzymes. We use these enzymes as a tool for our characterization of promoters.
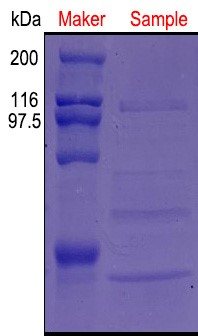
SDS-page detection qualitative analysis
Methods and results
Take 1 mL fermented liquid, in the 12000r∙min-1, 1 min the centrifugal, the fermented supernatant as samples to identify the Protein SDS-page electrophoresis of pullulan enzyme expression, using SDS-PAGE of concentrated gum concentration was 5%, the concentration of separation gel was 9%, take samples of 30 mu, 4 x L SDS-page Protein Loading Buffer 10 mu L and blending boil after 5 min, 5000 r, 2 min min-1 centrifugal, samples from 15 mu L processed Protein to join the fibrin glue lane, 5 μl high molecular weight protein Marker was added.
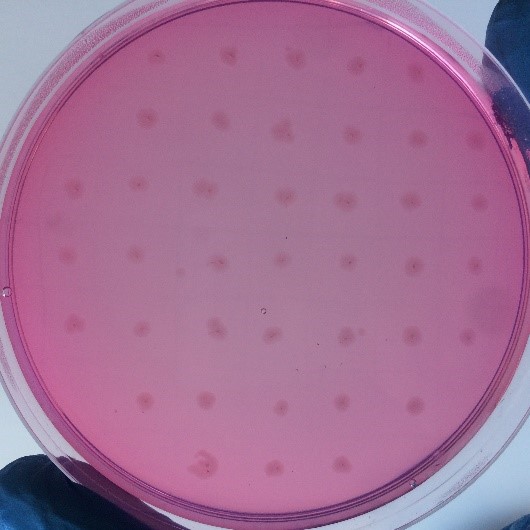
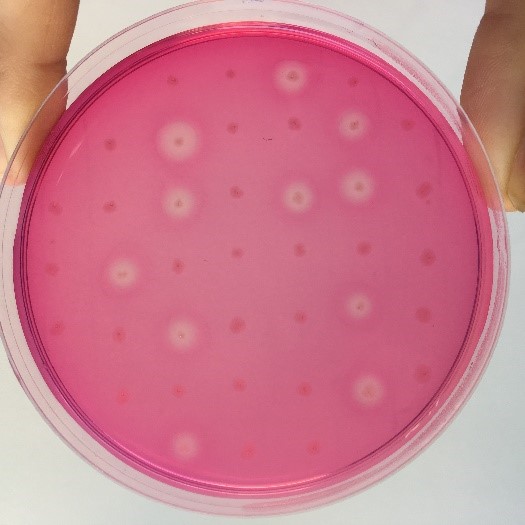
After the completion of the unique plasmid construction, we introduced the plasmid into Bacillus subtilis for strain construction and screening. In addition to the screening of antibiotics, we also did plate screening, adding 2% red Pullulan in the basic LB medium, and obtained the red medium for identification. In the medium of the recombinant strain, we can get the different phenomenon that the pullulanase express or not.
Figure 2:medium including red Pullulan without expression of pullulanase</p>
Figure 3:medium including red Pullulan with pullulanase expression</p>
Determination of enzyme activity: 1mL fermentation broth was centrifuged at 12000r∙ min-1 for 1min, and the supernatant of the obtained fermentation broth was diluted appropriately for the determination of extracellular enzyme activity. The determination method of pullulan enzyme activity was as follows:
First, a properly diluted fermentation supernatant of 200 microns was added to a tube with a plug scale in an ice bath, followed by a 2% substrate of puluan 200 microns prepared with a buffer of acetic acid-sodium acetate (pH4.5). The product was reacted in a 60℃ water bath for 20 minutes, and then immediately cooled in an ice water bath. The treatment of the control group was as follows: before the reaction at 60℃, only 200 mul of enzyme solution was added, and pululan substrate was not added. After the reaction, 200 mul of substrate was added in the ice bath. Then, 600 microns of DNS reagent was added to each cooled tube of reaction mixture and reacted in the boiling water bath for 5 minutes before being transferred to the ice water bath for rapid cooling. Finally, 3mL deionized water was added to each tube of reaction solution, and 200 microns were added to the porous cell culture plate after the mixture. The OD540 of each tube of reaction solution was determined by an enzyme marker. A unit of pullulanase activity is defined as the amount of enzyme required to release 1 mol of reducing sugar per minute under measured conditions. All enzyme activity is the average value of our three measurements, and the calculation formula of enzyme activity is as follows:
Enzyme activity (U· ml-1) = dilution × (reaction OD540- control OD540)/(0.0027×180×20×0.2)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |