Part:BBa_K2955002
glpB
glpB is a gene that was found to be related to degradation of glyphosate in the bacterium Burkholderia psuedomallei (formerly Psuedomonas). This gene was utilized as part of a two gene system along with glpA (BBa_K2955001) with the overall goal of degrading glyphosate. This gene was reported by Penaloza-Vazquez et al (1994) as allowing glyphosate to be used as a phosphorous source by breaking the C-N bond.
[1] Penaloza-Vazquez, A., Mena, G. L., Herrera-Estrella, L., & Bailey, A. M. (1995). Cloning and Sequencing of the Genes Involved in Glyphosate Utilization by Pseudomonas pseudomallei. APPL. ENVIRON. MICROBIOL., 61, 6
Usage and Biology
glpB was used as part of a two part system with the goal of increasing cell survivability and degradation of glyphosate. glpB is associated with the breakdown of glyphosate through the glyphosate oxidoreductase pathway, in which the glyphosate molecule is broken at the C-N bond to form aminomethylphosphonic acid (AMPA) and glyoxylate. In our system, glpB was regulated by pLac, and in experiments expression was induced by adding IPTG.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 147
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Methods
This part was tested by running growth curves under various conditions. Growth curve data was collected using a Bioscreener, Bioscreen C. Overnight cultures of the full plasmid, which had both glpA (BBa_K2955001) and glpB , and a control culture were grown overnight in tubes of LB+chloramphenicol at 37C and 220 RPM. 3.0 mL of the tested concentrations of pure glyphosate and Roundup were prepared with 3 uL of chloramphenicol. IPTG was added to reach a final concentration of 0.5 mM IPTG in induced samples.Each concentration was tested with 4 replicates. Each well was inoculated with 3 uL of cells and 300 uL of the stock concentrations. The plates were left in the Bioscreener for 96 hours at 37 C and meadium shaking. Measurements were taken every 30 minutes. It was not possible to test glpB independently of glpA due to the constitutive promoter regulating glpA. The following data is taken in comparison to Rounddown when glpB is not expressed.
Results
Compared to when only glpA was expressed, the induction of glpB seemed to improve growth in pure glyphosate. The following figure shows the growth in pure glyphosate when IPTG was added to compared to when it wasn't present. In the presence of IPTG, Rounddown was able to grow to a higher peak OD than when there was no IPTG added. This could be potentially be because the cells are able to utilize the glyphosate as a phosphate source. In the presence of IPTG the Rounddown also showed a much shorter lag phase, reaching peak OD at a much faster rate.
Figure 1
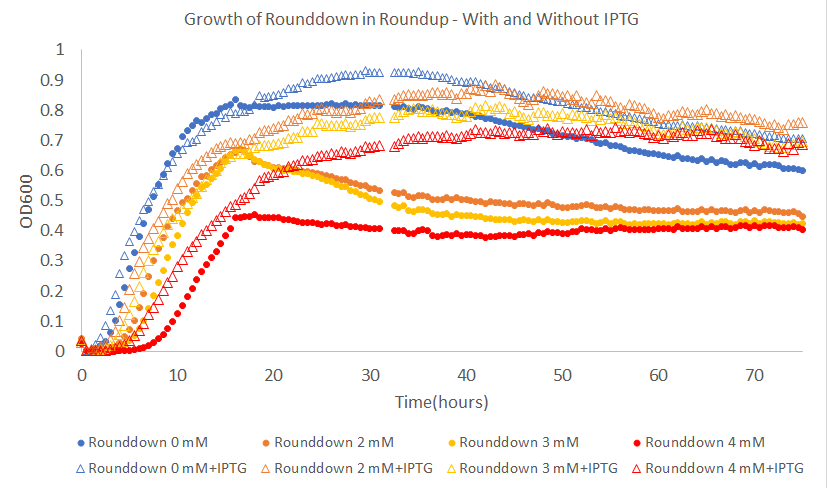
The main effect of glpB in a pure glyphosate system it to decrease the length of the lag phase. A slightly different effect is seen in a system utilizing Roundup. When IPTG is added to cells growing in Roundup, the peak OD reached by the cells is very similar to that reached when no Roundup is present. This is shown in the following figure. This increase in peak OD is indicative of increased tolerance towards the glyphosate and surfactant mixture than when glpA alone is expressed.
Figure 2
Burkholderia
| None |