Part:BBa_K2906001
sfGFP with N-Terminal secretion signal (OmpA)
Superfolder GFP also known as sfGFP is a GFP-derived green fluorescent protein. (Pédelacq et al., 2006). GFP is a protein isolated from the jellyfish Aequorea Victoria that exhibits green fluorescence when exposed to light in the blue to the ultraviolet range (Prendergast and Mann, 1978; Tsien, 1998). A series of mutations performed by Pédelacq et al. obtained a GFP variant able to rapidly fold and mature. This ultimately also leads to enhanced fluorescence intensity (Frenzel et al., 2018). This composite part is made of sfGFP (BBa_K1321337) with a hydrophobic tag BBa_K2906007 and an N-Terminal secretion signal (OmpA) BBa_K208003 to secrete the protein into the cytoplasm. It is expressed by a pTet promoter BBa_R0040. Below we discuss our reasoning behind these choices and our quantitative and qualitative findings. Unfortunately, this part only expressed limited colour and was unable to be secreted into the media.
Contents
OmpA signal peptide:
Outer membrane peptide A (OmpA) is a conserved protein domain found at the outer membrane of many gram-negative bacteria such as E. coli (Wang, 2002; Smith et al., 2007). The OmpA secretion signal is located in the N-terminal region of the protein of interest and becomes trafficked through a type II-dependent secretion system (Kotzsch et al., 2011). The signal peptide is cleaved by the secretion machinery in the plasma membrane of the bacterium. The resulting protein is secreted and released in an active mature form. This secretion signal peptide was therefore selected due to its reposted high efficiency of secretion during high protein expression (Pechsrichuang et al., 2016).
Hydrophobic Tag:
Since we aimed at the creation of new hair dyes that would not damage the cortex of hair, we did not want our designed coloured protein-based dyes to infiltrate the cortex as this will lead to cuticle opening and weaken the hair itself. Therefore, both of our secreted variants also contained a novel hydrophobic tag BBa_K2906007
Characterisation:
To show whether this part worked, we decided to measure fluorescence, normalised by OD. Optical Density values were measured at 600 nm. We grew the cells to an OD600 of ~0.6 and induced them with 100 nM of anhydrotetracycline. Then, they were loaded in triplicates into a 96-well plate and left overnight. For OD values, the blank was LB media, and for RFU the blank was E. coli TOP10 since it does not express any colour. The values were individually normalised by dividing RFU/OD and then averaged to plot the mean against time. An RFU value of 0 corresponds to baseline E. coli TOP10 measurements.
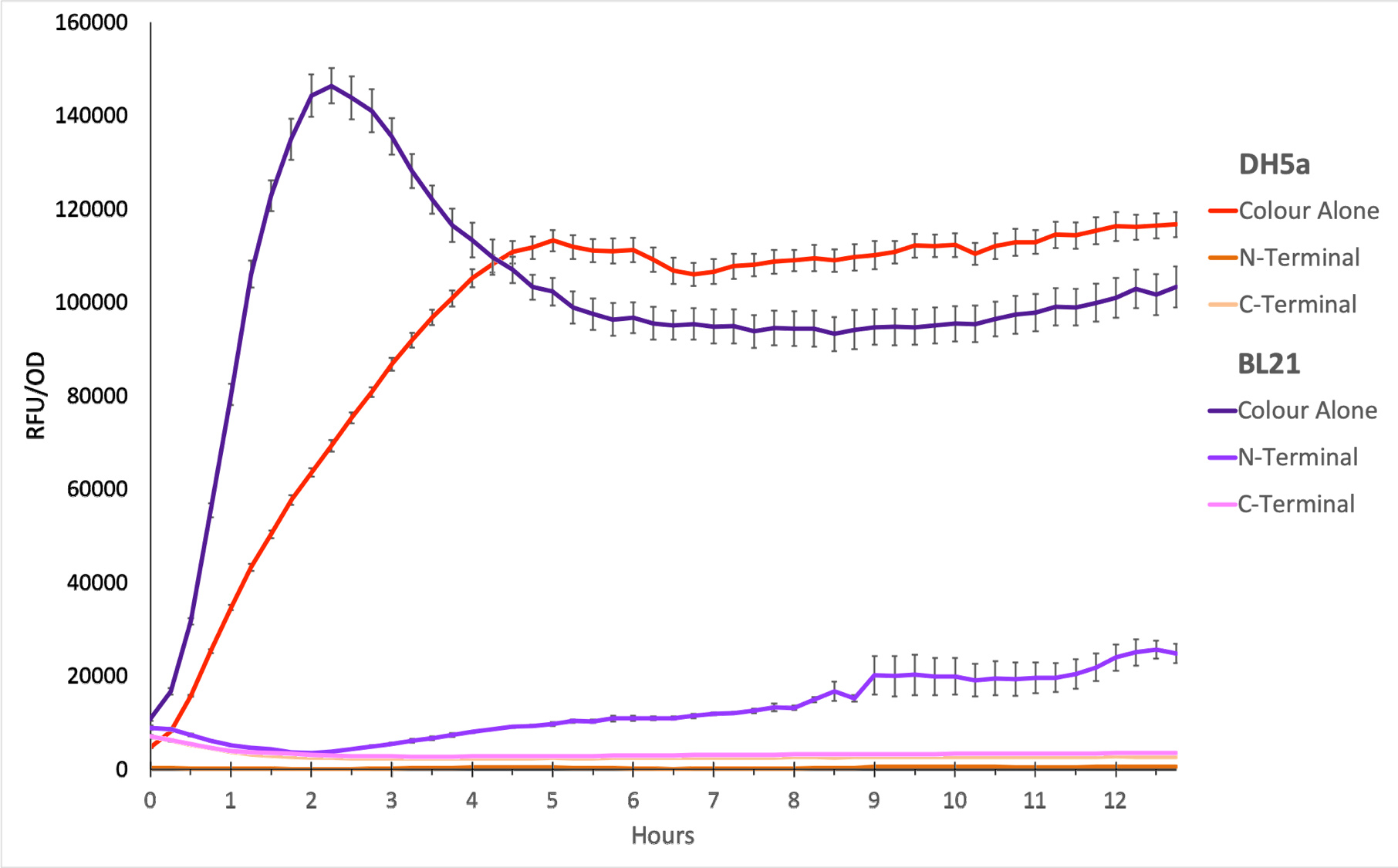
Figure 1. The plot shows the mean RFU/OD from three replicates of each construct expressed in E. coli DH5⍺ and BL21(DE3). The OD was measured at 600 nm and GFP fluorescence was measured at Ex ƛ 485, Em ƛ 510, every 15 minutes for 13 hours. The RFU values were normalised by the OD and the triplicates averaged. All values have been blank-corrected. A total of 52 recordings were made per well, with three wells per construct. The N-Terminal construct shows some fluorescence in BL21 cells, but none in DH5a.
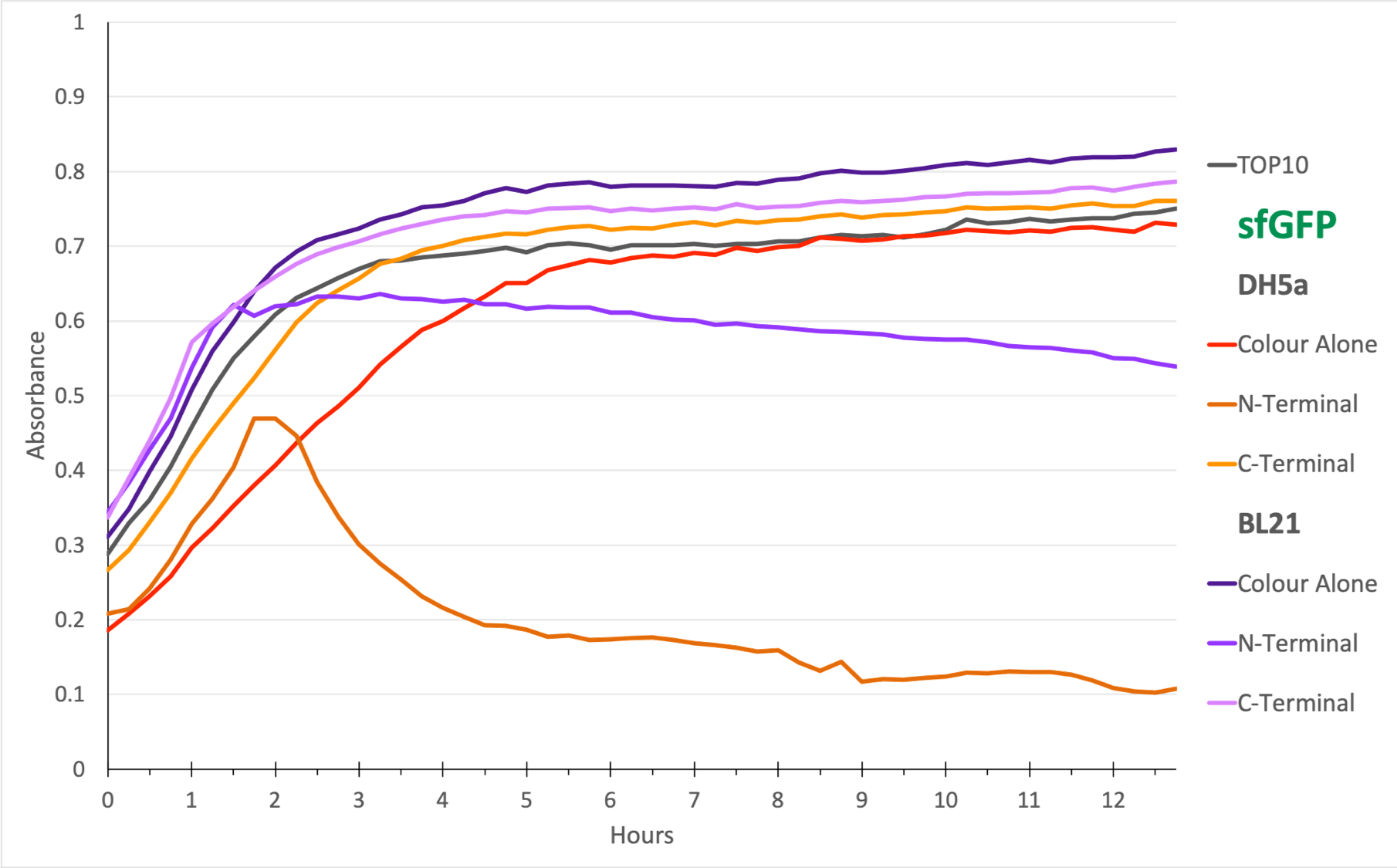
Figure 2. The N-terminal transformed bacteria seem to decline over time. This is true in E. coli DH5⍺ and BL21(DE3) and suggests a degree of stunted growth. From the growth curve, we can conclude that sfGFP with OmpA N-terminal must be causing toxic aggregates in the E. coli cells. TOP10 was the negative control since it is E. coli that does not express any colour.
Qualitative Data:
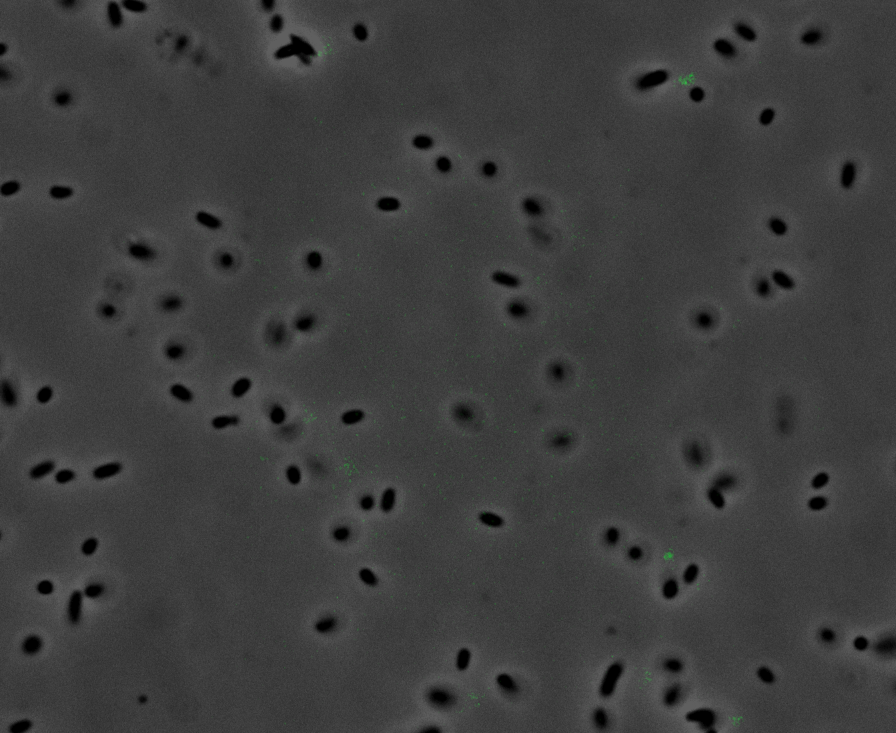
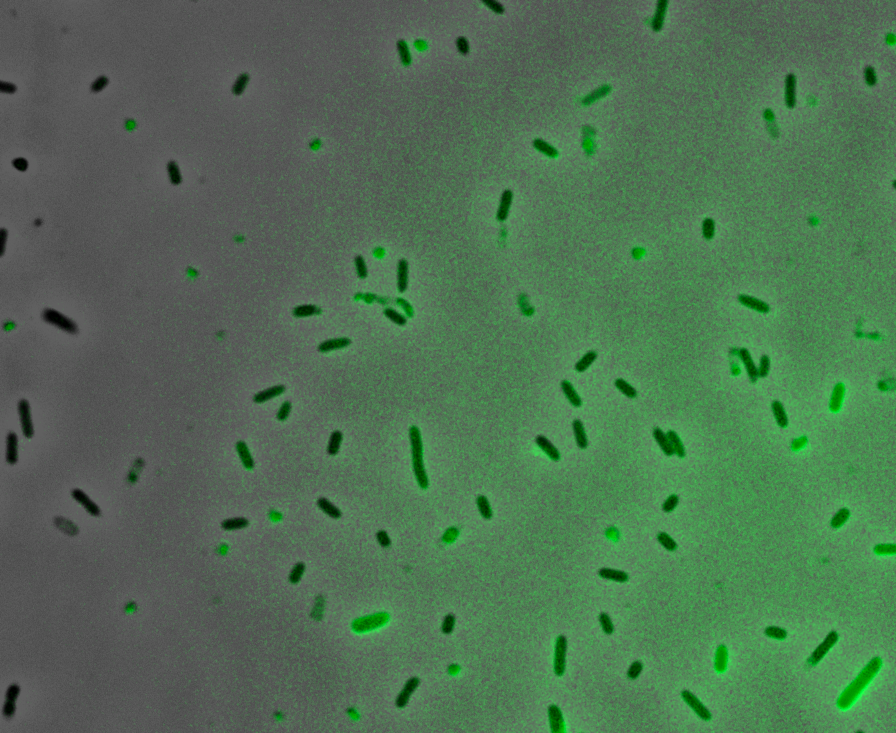
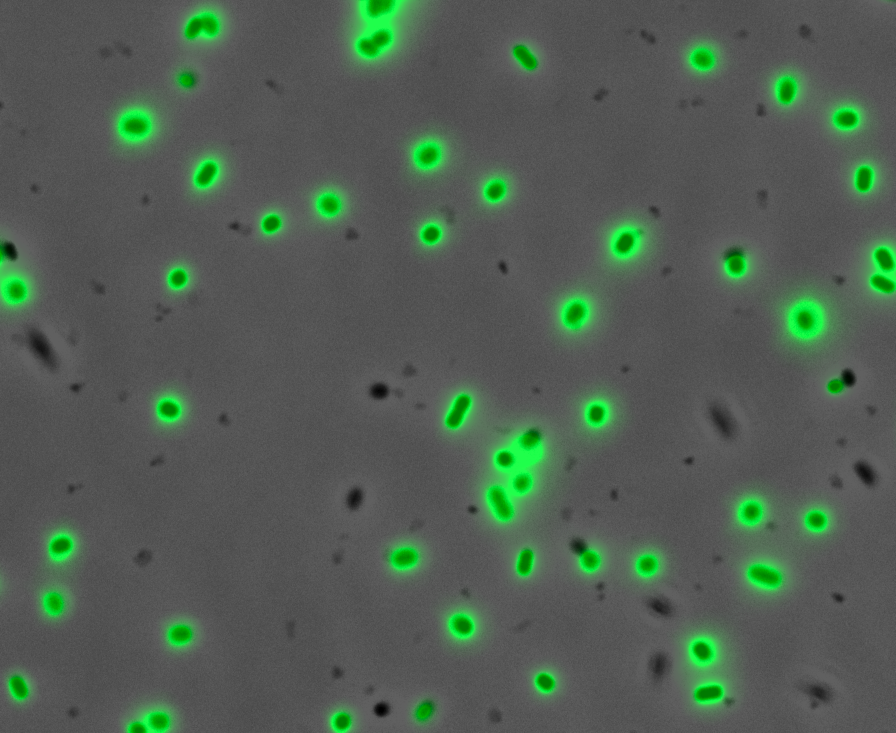
Fluorescence Microscopy: After quantifying the data for sfGFP we decided to perform fluorescence microscopy. We did this since the experiments above show that there is a limited amount of sfGFP detected through fluorescence spectrometry, but no colour was seen. This would allow us to see the percentage of bacteria in a sample producing colour, if any, as well as some phenotypic characteristics of the bacteria with the construct. Below you can see three images, the negative control, which was E. coli TOP10, showing very limited background GFP fluorescence, sfGFP with OmpA (BBa_K2906001) showing limited amounts of fluorescence, and BBa_K2906000, which clearly expresses sfGFP.
Figure 3. Fluorescent microscopy overlaid images of phase contrast and GFP filter. a) Negative control (E. coli TOP10) showing very slight background readings for fluorescence. b) sfGFP + OmpA (BBa_K2906001) showing very limited fluorescence in some bacteria and high background noise, this shows that fluorescence is not intense. c) BBa_K2906000 showing appropriate levels of fluorescence in most bacteria. The images above are composite overlaid images made from phase contrast and GFP-filter captures. They were processed using ImageJ.
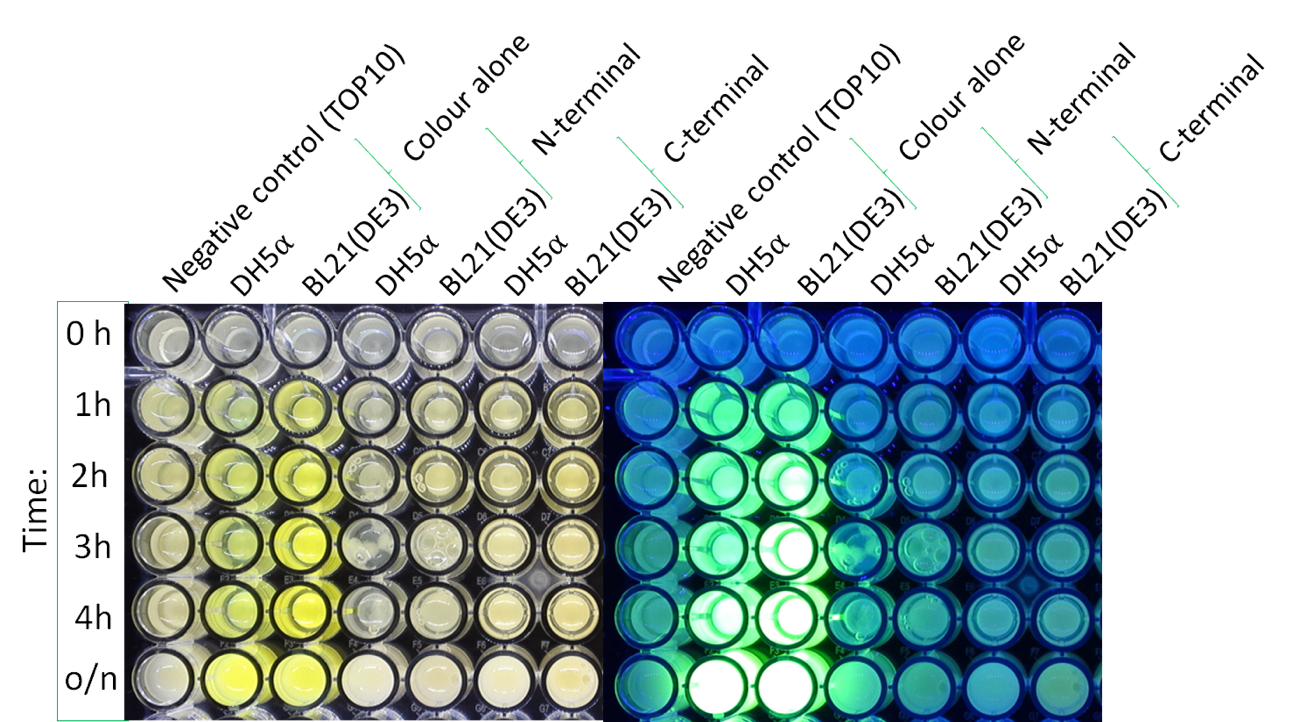
Colour:
The image below shows the visible colour of the pellet obtained under normal light and UV light. This was done at five different time points after induction with 100 nM of anhydrotetracycline. sfGFP with OmpA is labelled ''N-Terminal'' and can be seen present in both DH5a and BL21.
References:
Frenzel, E., Legebeke, J., Van Stralen, A., Van Kranenburg, R. and Kuipers, O. P. (2018) ‘In vivo selection of sfGFP variants with improved and reliable functionality in industrially important thermophilic bacteria’, Biotechnology for Biofuels. BioMed Central Ltd., 11(1). doi: 10.1186/s13068-017-1008-5.
Kotzsch, A., Vernet, E., Hammarström, M., Berthelsen, J., Weigelt, J., Gräslund, S. and Sundström, M. (2011) ‘A secretory system for bacterial production of high-profile protein targets’, Protein Science, 20(3), pp. 597–609. doi: 10.1002/pro.593.
Pechsrichuang, P., Songsiriritthigul, C., Haltrich, D., Roytrakul, S., Namvijtr, P., Bonaparte, N. and Yamabhai, M. (2016) ‘OmpA signal peptide leads to heterogenous secretion of B. subtilis chitosanase enzyme from E. coli expression system’, SpringerPlus. SpringerOpen, 5(1). doi: 10.1186/s40064-016-2893-y.
Pédelacq, J. D., Cabantous, S., Tran, T., Terwilliger, T. C. and Waldo, G. S. (2006) ‘Engineering and characterization of a superfolder green fluorescent protein’, Nature Biotechnology, 24(1), pp. 79–88. doi: 10.1038/nbt1172.
Prendergast, F. G. and Mann, K. G. (1978) ‘Chemical and Physical Properties of Aequorin and the Green Fluorescent Protein Isolated from Aequorea forskålea†’, Biochemistry, 17(17), pp. 3448–3453. doi: 10.1021/bi00610a004.
Smith, S. G. J., Mahon, V., Lambert, M. A. and Fagan, R. P. (2007) ‘A molecular Swiss army knife: OmpA structure, function and expression.’, FEMS microbiology letters, 273(1), pp. 1–11. doi: 10.1111/j.1574-6968.2007.00778.x.
Tsien, R. Y. (1998) ‘THE GREEN FLUORESCENT PROTEIN’, Annual Review of Biochemistry. Annual Reviews, 67(1), pp. 509–544. doi: 10.1146/annurev.biochem.67.1.509.
Wang, Y. (2002) ‘The function of OmpA in Escherichia coli’, Biochemical and Biophysical Research Communications, 292(2), pp. 396–401. doi: 10.1006/bbrc.2002.6657.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 184
| None |