Part:BBa_K2586019
GAT: Glyphosate N-Acetyltransferase
This part encodes for an enzyme that n-acetylates the herbicide and antibiotic glyphosate.
The GAT-enzyme is able to confer glyphosate resistance to organisms such as Escherichia coli, Arabidopsis, tobacco and maize.
The acetylation of glyphosate may provide a new and alternative strategy in glyphosate support on crop plants.
We integrated this part into soil bacterium Bacillus subtilis to enable the bacteria to inactivate glyphosate, which would otherwise be deadly to them.
| Molecular weight | 17 kDa |
| Protein length | 146 aa |
| Gene length | 483 bp |
| Function | mediates glyphosate tolerance |
| Product | N-acetyl glyphosate |
| Ideal pH | 6.8 |
| Essential | no |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
Engineering bacteria to disarm glyphosate
Many years ago it has been found that glyphosate can be inactivated by acetylation yielding N-acetylglyphosate ( Castle et al.,2004). Because the acetylated form of glyphosate has low affinity for the active site of EPSP synthase, it is nonherbicidal. The glyphosate N-acetyltransferase GAT from the Gram-positive soil bacterium Bacillus licheniformis transfers the acetyl group from acetyl-CoA onto the amino group of the weedkiller ( Castle et al.,2004).
Interestingly, B. subtilis possesses a putative glyphosate N-acetyltransferase, which is designated as YitI and shares 59% overall amino acid identity with GAT of B. licheniformis. However, the native GAT enzymes are very poor catalysts for N-acetylation of glyphosate ( Castle et al.,2004). Moreover, despite extensive screening of biological amines, including amino acids, nucleotides and antibiotics, the physiological substrates for the native enzymes are unknown (Siehl et al., 2007 JBC). As an alternative strategy for glyphosate resistance involving enzymatic conversion of glyphosate to N-acetylglyphosate the GAT was also subjected to directed evolution for creating an enzyme with higher efficiency and increased specificity for the herbicide ( Castle et al.,2004). When introduced into plants, optimized gat genes confer robust tolerance to glyphosate ( Castle et al.,2004). Therefore, we expect that bacteria like B. subtilis expressing the optimized GAT should tolerate high amounts of glyphosate even though it is taken up from the environment via the high-affinity glyphosate transporter GltT (see above). To evaluate the potential of the GAT to allow B. subtilis in the presence of glyphosate concentrations that are toxic for the wild type bacteria, we wanted to express the gat gene in B. subtilis (Figure 1).
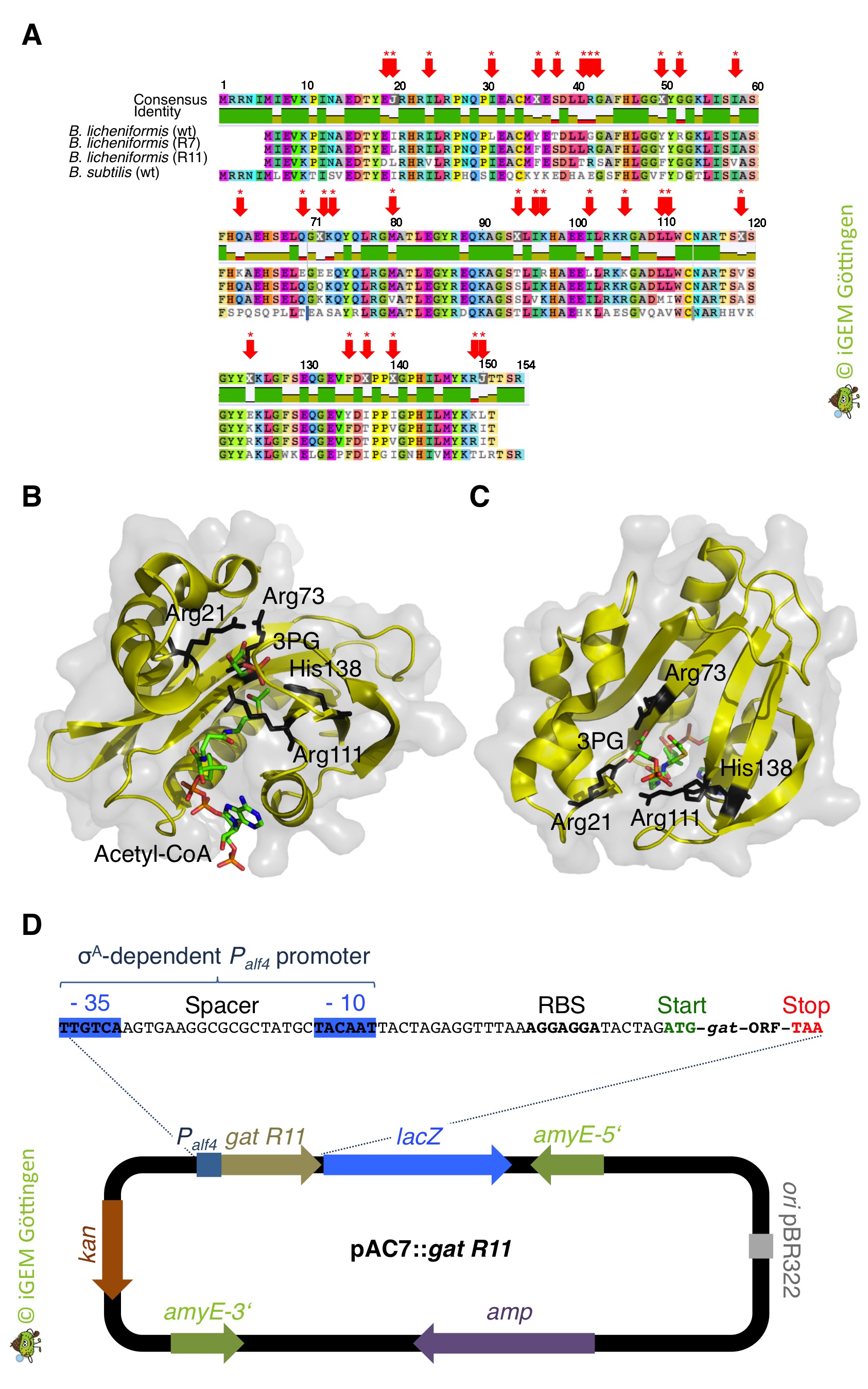
GAT confers tolerance to glyphosate
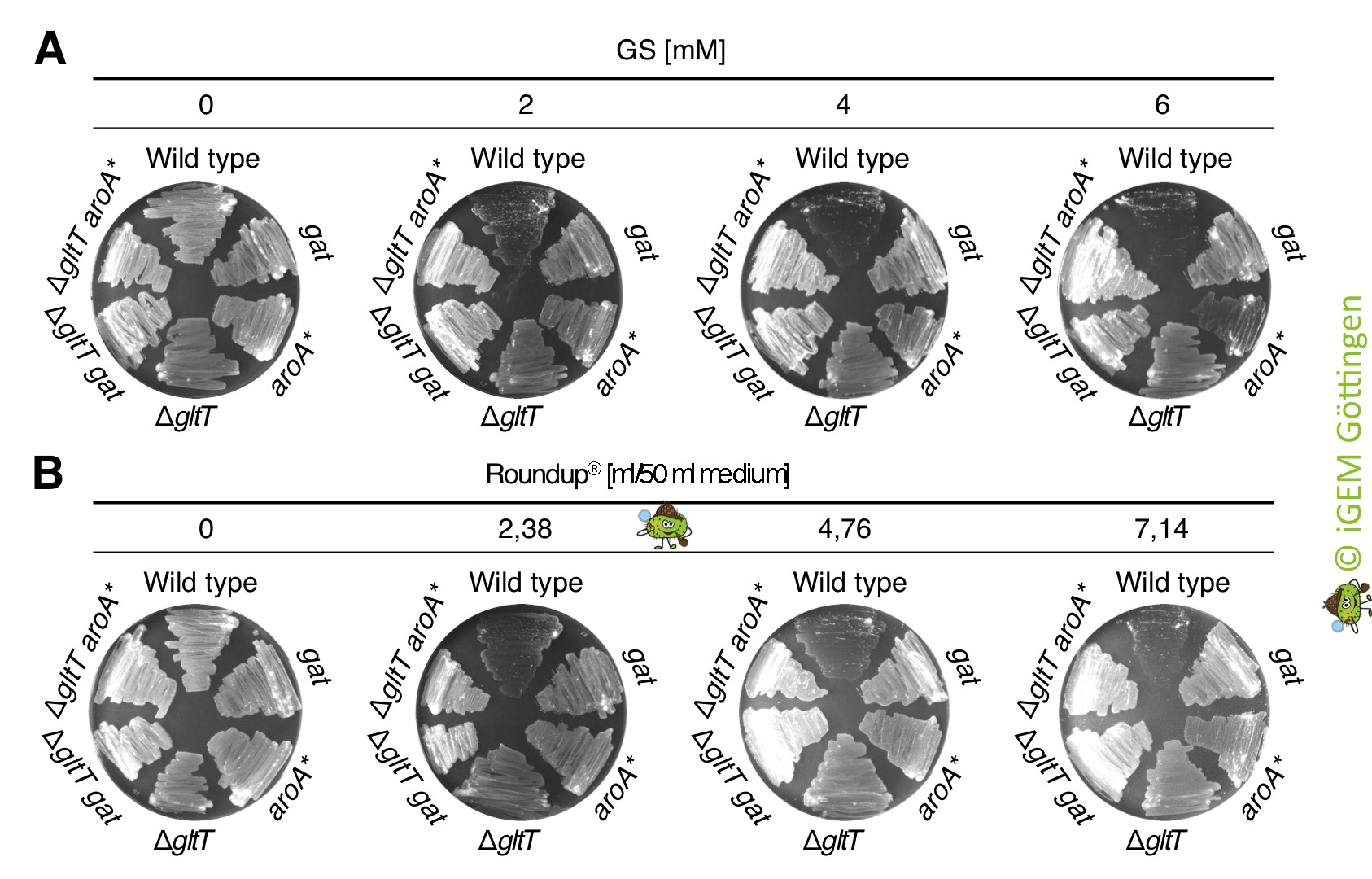
We ordered the gat gene as a g-Block from Integrated DNA Technologies, which was free of charge because the company supports the iGEM competition. Next, we amplified the gat gene by PCR using the g-Block as template DNA. An artificial promoter and a ribosome binding site was attached during the PCR (Figure 1). The promoter-gene fusion was cloned and introduced in single copy into the genomes of the B. subtilis wild type strain and the gltT mutant strain to further increase its glyphosate tolerance (Figure 2). To assess the potential of the GAT to disarm glyphosate in the different B. subtilis strains, we propagated the bacteria on CS-Glc minimal medium agar plates that were supplemented with different amounts of glyphosate. As shown in Figure 2, the strains synthesizing the GAT tolerated significantly more glyphosate than the wild type strain. Moreover, the inactivation of the gltT glyphosate transporter gene and the overexpression of the gat gene confers high-level glyphosate tolerance to the bacteria. Thus, when introduced into B. subtilis , the optimized gat gene confers indeed robust tolerance to glyphosate. To conclude, the engineered bacteria possess two interesting properties: (i) B. subtilis promotes plant growth by protecting plants against pathogens and (ii), the bacteria take up glyphosate from the environment and inactivate the weedkiller!
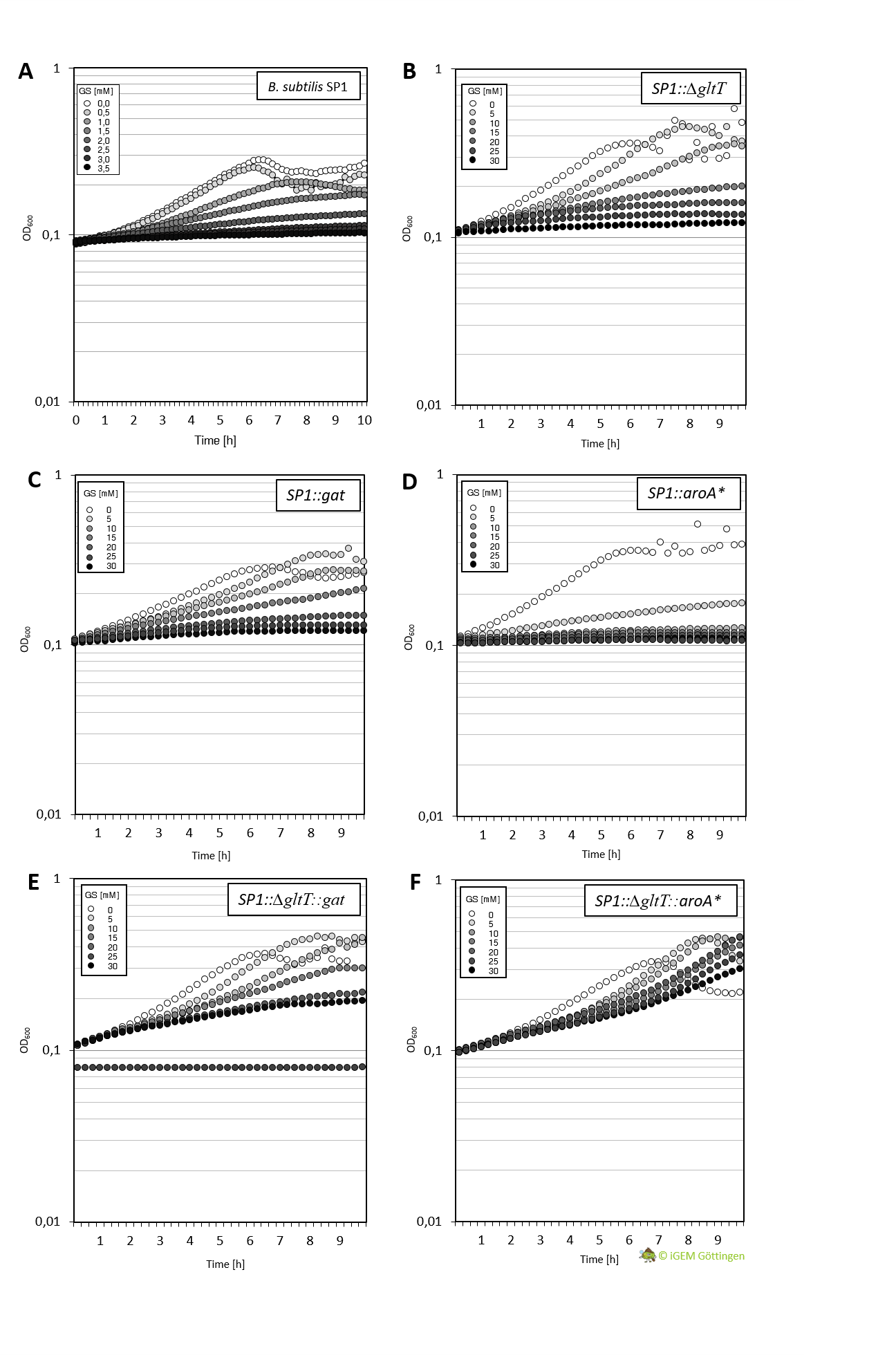
Characeterization of GAT conferred tolerance to glyphosate
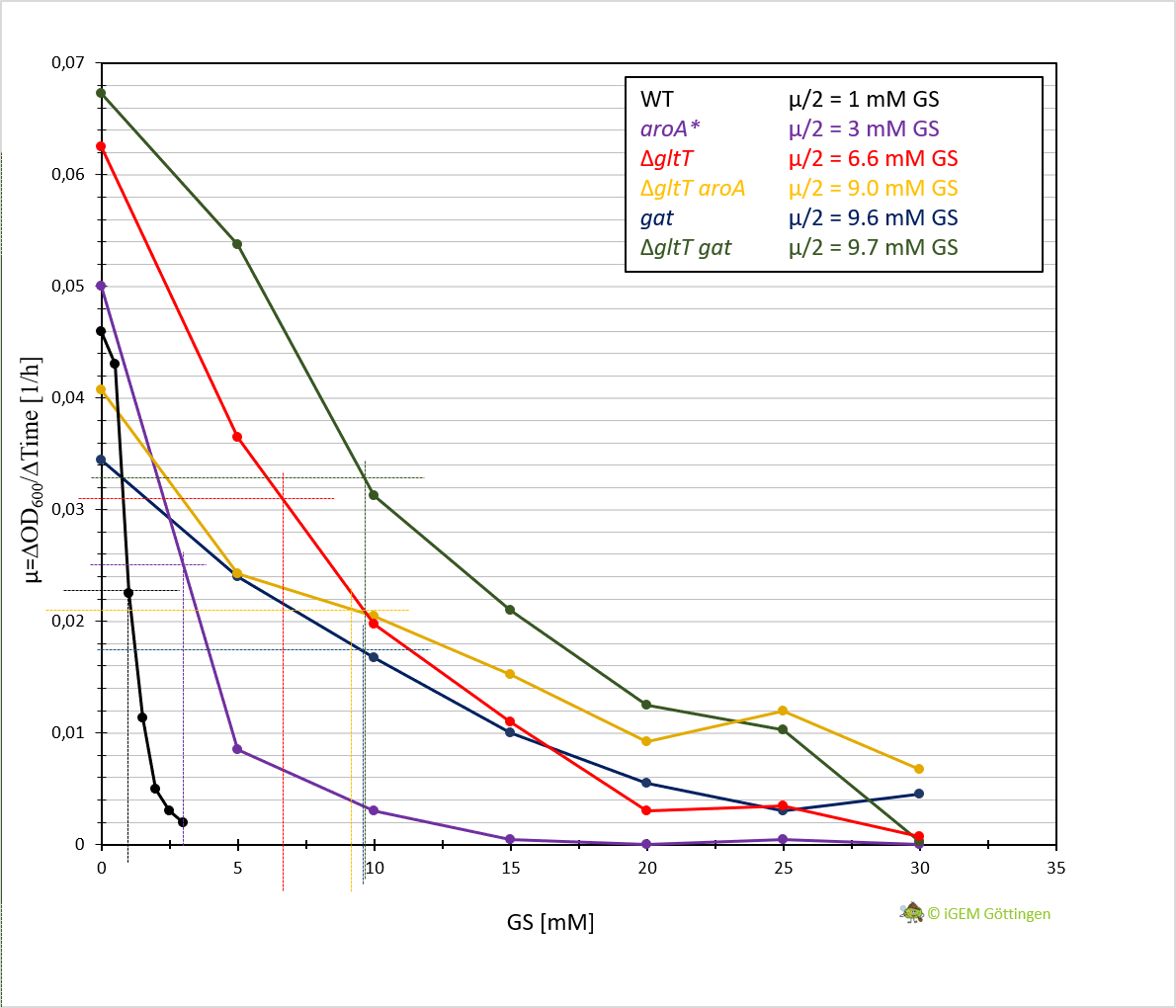
Cultivation of the different strains in liquid media allows more detailed characterisation. We analysed growth of the bacteria in CS-Glu medium supplemented with increasing amounts of glyphosate, using a platereader.
In general, the analysis supports previous results. The strain containing aroA* confers only a slight increase in resistance in comparison to the WT strain and hence a significantly lower resistance in comparison the ∆gltT strain (Figure 3 A, B and D ). However, for the strain combining aroA* and ∆gltT a 9-fold higher glyphosate concentration is needed to reduce the growth rate by 50% (Figure 3 F. Figure Y ).
Further characterisation of the Gat enzyme supports previous results and underline significant increased resistance in both strains conferring gat or a combination of gat and ∆gltT (Figure 3 C and E, Figure 4).
A novel nechanism conferring resistance to glyphosate
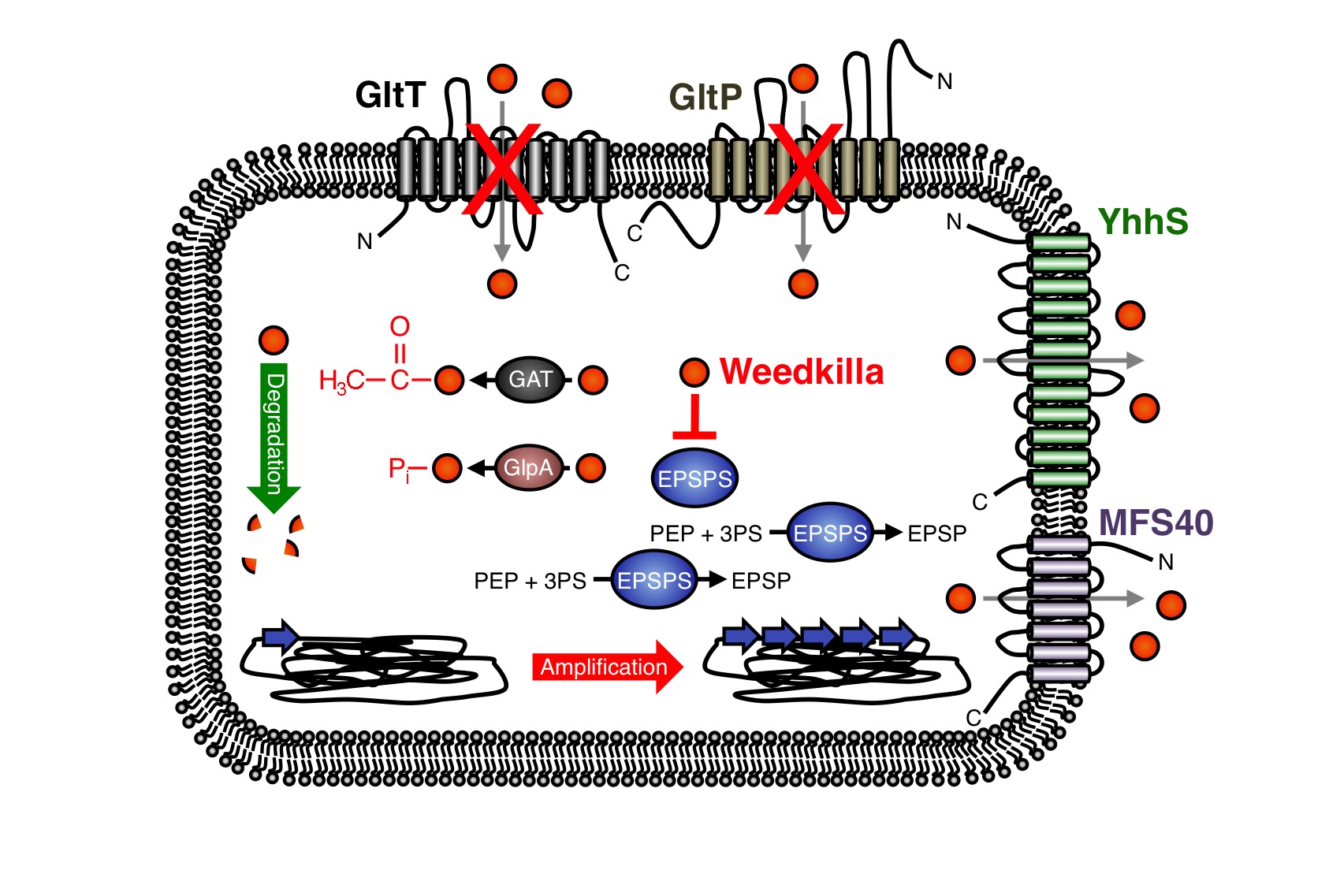
In the past years, the underlying molecular mechanisms conferring resistance to glyphosate have been intensively studied in bacteria and plants. In many cases glyphosate resistance is directly linked to the target of the herbicide. For instance, bacteria like S. aureus are a priori resistant to glyphosate because this organism synthesizes an insensitive EPSP synthase (13-16). Moreover, increased cellular levels of the EPSP synthase, which can be achieved either by overexpression of the coding gene or by gene amplification, can confer resistance to glyphosate (Figure 5) (9, 17-21). Due to the increased cellular levels of the EPSP synthase, the glyphosate is probably titrated away and sufficient amounts of the precursor for aromatic amino acid biosynthesis can be produced.
As mentioned in the design section, several glyphosate-insensitive EPSP synthase mutant variants have been isolated and engineered (11, 22-35). Often, a single amino acid exchange is sufficient to render the EPSP synthase insensitive to glyphosate. However, although glyphosate resistance is frequently linked to the target of the herbicide, resistance against the herbicide may occur by other means. Several studies have demonstrated that glyphosate can be detoxified by covalent modification. For instance, the Gat glyphosate N-acetyltransferase from Bacillus licheniformis, which was subjected to directed evolution for creating an enzyme with higher efficiency and increased specificity for the herbicide, converts glyphosate to N-acetylglyphosate, which is not herbicidal and is not an effective inhibitor of EPSP synthases (Figure 9) (36-39). Moreover, the hygromycin phosphotransferases Hph and GlpA from E. coli and Pseudomonas pseudomallei, respectively, phosphorylate glyphosate and thus confer tolerance to the herbicide (Fig. 3) (40,41). Interestingly, the gat gene has also been used as a selection marker for genetic engineering of bacteria (39). The enzymes that covalently modify glyphosate have been successfully introduced into crops to increase herbicide resistance (42).
Recently, it has been also demonstrated that enhanced export of glyphosate can reduce toxicity of the herbicide. For instance, the overexpression of the uncharacterized membrane proteins MFS40 and YhhS from Aspergillus oryzae and E. coli, respectively, with similarity to the major facilitator secondary transporter superfamily, enhance glyphosate tolerance of E. coli (Figure 5) (43,44). It will be interesting to test whether these transporters are suitable for engineering crops to enhance glyphosate tolerance. Proteins of unknown function can also increase glyphosate tolerance. For instance, the overexpression of the igrA gene product from the Pseudomonas sp. strain PG2982 increases glyphosate tolerance in transgenic rice (45-47). Finally, many bacteria can survive in the presence of glyphosate because they are able to degrade the herbicide (Figure 5) (48-57). To conclude, several mechanisms of glyphosate resistance have been described over the past years and novel mechanisms allowing survival with the herbicide are certainly identified in the future. We have observed that the deletion of the gltT gene in B. subtilis confers high-level glyphosate resistance to the bacteria. The B. subtilis gltT mutant strain could be a suitable host to identify glyphosate uptake systems from plants by expressing cDNA libraries and by screening for transformants that are unable to grow with glyphosate.
References
1. Fischer et al. (1986) J. Bacteriol. 168: 1147-1154
2. Zeigler et al. (2008) J. Bacteriol. 190: 6983-6995.
3. Barbe et al. (2009) Microbiology 155: 1758-1775
4. Kearse et al. (2012) Bioinformatics 28: 1647-1649.
5. Omasits et al. (2014) Bioinformatics. 30: 884-886.
6. Michna et al. (2014) Nucleic Acids Res. 42: D692–D698.
7. Zaprasis et al. (2015) Appl. Environ. Microbiol. 81: 250-259.
8. Tolner et al. (1995) J. Bacteriol. 177: 2863-2869.
9. Amrhein et al. (1980) Naturwissenschaften 67: 356-357.
10. Comai et al. (1983) Science 221: 370-371.
11. Stalker et al. (1985) J. Biol. Chem. 260: 4724-4728.
12. Gundlach et al. (2017) Sci. Signal. 10: eaal3011.
13. Priestmann et al., (2005) FEBS Lett. 579: 728-732.
14. Cao et al. (2012) PLoS One. 7: e38718.
15. Light et al. (2016) Biochemistry 55: 1239-1245.
16. Liu & Cao, (2018) Biotechnol Lett. 40: 855-864.
17. Rogers et al. (1983) Appl. Envrion. Microbiol. 46: 37-43.
18. Gaines et al. (2011) J Agric Food Chem. 59: 5886-5889.
19. Juglam et al. (2014) Plant Physiol. 166: 1200-1207.
20. Sammons & Gaines (2014) Pest. Manag. Sci. 70: 1367-1377.
21. Dillon et al. (2017) Plant Physiol. 173: 1226-1234.
22. Sost & Amrhein (1990) Arch. Biochem. Biophys. 282: 433-436.
23. Padgette et al. (1991) J. Biol. Chem. 266: 22364-22369.
24. He et al. (2001) Biochim. Biophys. Acta 1568: 1-6.
25. He et al. (2003) Biosci. Biotechnol. Biochem. 67: 1405-1409.
26. Bearson et al. (2002) Plant Physiol. 129: 1265-1275.
27. Eschenburg et al. (2002) Planta 216: 129-135.
28. Sun et al. (2005) Appl. Envrion. Microbiol. 71: 4771-4776.
29. Zhou et al. (2006) Plant Physiol. 140: 184-195.
30. Healy-Fried et al. (2007) J. Biol. Chem. 282: 32949-32955.
31. Vande Berg et al. (2008) Pest. Manag. Sci. 64: 340-345.
32. Funke et al. (2009) J. Biol. Chem. 284: 9854-9860.
33. Tian et al. (2010) Appl. Environ. Microbiol. 76: 6001-6005.
34. Pollegioni et al. (2011) FEBS J. 278: 2753-2766.
35. Chekan et al. (2016) MedChemComm. 7: 28-36.
36. Castle et al. (2004) Science 304: 1151-1154.
37. Siehl et al. (2005) Pest. Manag. Sci. 61: 235-240.
38. Siehl et al. (2007) J. Biol. Chem. 282: 11446-11455.
39. Norris et al. (2009) Appl. Environ. Microbiol. 75: 6062-6075.
40. Rao et al. (1983) Antimicrob. Agents Chemother. 24: 689-695.
41. Penaloza-Vazquez et al. (1995) Appl. Environ. Microbiol. 61: 538-543.
42. Liu et al. (2015) Transgenic Res. 24: 753-763.
43. Staub et al. (2012) J. Ind. Microbiol. Biotechnol. 39: 641-647.
44. Tao et al. (2017) Pestic. Biochem. Physiol. 140: 65-68.
45. Fitzgibbon & Braymer, (1988) Appl. Environ. Microbiol. 54: 1886-1888.
46. Fitzgibbon & Braymer, (1990) Appl. Environ. Microbiol. 56: 3382-3388.
47. Fartyal et al. (2018) Front. Plant. Sci. 13: 144.
48. Pipke et al. (1980) Appl. Environ. Microbiol. 53: 974-978.
49. Shinabarger et al. (1986) J. Bacteriol. 168: 702-707.
50. Liu et al. (1991) Appl. Environ. Microbiol. 57: 1799-1804.
51. Dick & Quinn, (1995) Appl. Microbiol. Biotechnol. 43: 545-550.
52. Singh & Walker, (2006) FEMS Microbiol. Rev. 30: 428-471.
53. Castro et al. (2007) J. Environ. Sci. Health B. 42: 883-886.
54. Hove-Jensen et al. (2014) Microbiol. Mol. Biol. Rev. 78: 176-197.
55. Kryuchkova et al. (2014) Microbiol. Res. 169: 99-105.
56. Sviridov et al., (2011) Biochemistry (Mosc). 76: 720-725.
57. Sviridov et al. (2015) Prikl. Biokhim. Mikrobiol. 51: 183-190.
| protein |