Part:BBa_K2323001:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K2323001
Lethbridge_HS iGEM 2019
Both parts of our system rely on the protein Cas13a, so it was imperative that we purify the protein for future use in our Cas13a activity assay. Our team successfully purified Lbu Cas13a protein while we were unable to purify Lwa Cas13a to a high enough concentration through Nickel Affinity Chromatography and Size Exclusion Chromatography. Initially, our team tried to express Cas13a in E. coli BL21 DE3; however, we had issues with overexpression so we tried again using E. coli Rosetta DE3 cells and saw greater success (Figure 1).
 Figure 1. 10% SDS PAGE of overexpression of Lbu and Lwa Cas13a. Run at 180 V for 25 minutes and 200 V for 2.5 hours. Left to right: lane 1: Lbu before induction; lane 2: Lbu after induction; lane 3: Lbu before induction; lane 4: Lbu after induction; lane 5: Lwa before induction; lane 6: Lwa after induction; lane 7: Lwa before induction; lane 8: Lwa after induction; lane 9: 10-250 kDa ladder; lane 10: empty.
Figure 1. 10% SDS PAGE of overexpression of Lbu and Lwa Cas13a. Run at 180 V for 25 minutes and 200 V for 2.5 hours. Left to right: lane 1: Lbu before induction; lane 2: Lbu after induction; lane 3: Lbu before induction; lane 4: Lbu after induction; lane 5: Lwa before induction; lane 6: Lwa after induction; lane 7: Lwa before induction; lane 8: Lwa after induction; lane 9: 10-250 kDa ladder; lane 10: empty.
Those cells were lysed then centrifuged to separate the supernatant and cell pellet. The lysate was then introduced to a Nickel Sepharose affinity column to isolate the Cas13a protein. It was bound to the Nickel Sepharose due to the histidine tag. Please note that while Lwa Cas13a does not appear well on the scanned gel, a faint band appeared in the wash and elution fractions.
 Figure 2. 10% SDS PAGE of Lwa Cas13a protein after his tag purification. Left to right: lane 1: Lwa Lysate; lane 2: Lwa lysate post-binding; lane 3: Wash (pooled); lane 4: Elution 1; lane 5: elution 2; lane 6: elution 3; lane 7: elution 4; lane 8: elution 5; lane 9: 10-250 kDa Ladder; lane 10: empty.
Figure 2. 10% SDS PAGE of Lwa Cas13a protein after his tag purification. Left to right: lane 1: Lwa Lysate; lane 2: Lwa lysate post-binding; lane 3: Wash (pooled); lane 4: Elution 1; lane 5: elution 2; lane 6: elution 3; lane 7: elution 4; lane 8: elution 5; lane 9: 10-250 kDa Ladder; lane 10: empty.
To further purify Cas13a, we ran the partially purified protein solution through a column, that contained beads. The beads have small crevices and this causes proteins of different sizes to pass through during different times. Again, the band for Lwa did not show up well after scanning the gel.
 Figure 3. Purification of Lwa Cas13a using size exclusion chromatography. (A) 10% SDS PAGE of Lwa after size exclusion purification. Left to right: lane 1: Lwa concentrated post-affinity; lane 2: Lwa frac B7; lane 3: Lwa frac B6; lane 4: Lwa frac H5; lane 5: Lwa frac H3; lane 6: Lwa frac H1; lane 7: Lwa frac I1; lane 8: Lwa I7; lane 9: 10-250 kDa ladder; lane 10: Lwa frac I8. (B) Chromatogram generated by AktaPrime of Lwa Cas13a purified using a large Superdex75 column (GE Life Sciences).
Figure 3. Purification of Lwa Cas13a using size exclusion chromatography. (A) 10% SDS PAGE of Lwa after size exclusion purification. Left to right: lane 1: Lwa concentrated post-affinity; lane 2: Lwa frac B7; lane 3: Lwa frac B6; lane 4: Lwa frac H5; lane 5: Lwa frac H3; lane 6: Lwa frac H1; lane 7: Lwa frac I1; lane 8: Lwa I7; lane 9: 10-250 kDa ladder; lane 10: Lwa frac I8. (B) Chromatogram generated by AktaPrime of Lwa Cas13a purified using a large Superdex75 column (GE Life Sciences).
We noticed a large peak at absorbance 254 nm, which is the level at which nucleic acids absorbs. Since Cas13a binds and interacts with RNA this is likely what is causing the peak. We ran the sample on a urea page to confirm the presence of RNA (Figure 4). Members of the Wieden lab at the University of Lethbridge have worked with proteins that interact with RNA. To minimize these interactions, they purify this protein using anion exchange chromatography (Q Sepharose). We noticed that previous iGEM teams had not done this purification step in the past. To minimize any issues with other interacting RNAs, we recommend future iGEM teams complete this purification step in between nickel affinity chromatography and size exclusion chromatography. Due to time constraints, we were unable to test this purification methodology to see if it impacted our results.
 Figure 4. 10% Urea PAGE of Lbu and Lwa protein samples after affinity and size exclusion chromatography purification. Left to right: lane 1: High Range RiboRuler; lane 2: Lwa sample post-concentration after affinity purification; lane 3: Lbu sample post-concentration after size exclusion chromatography; lane 4: Lbu sample post-concentration after affinity chromatography.
Figure 4. 10% Urea PAGE of Lbu and Lwa protein samples after affinity and size exclusion chromatography purification. Left to right: lane 1: High Range RiboRuler; lane 2: Lwa sample post-concentration after affinity purification; lane 3: Lbu sample post-concentration after size exclusion chromatography; lane 4: Lbu sample post-concentration after affinity chromatography.
Initially, our team had planned on doing a triple plasmid transformation of our target fluorescent protein (GFP), crRNA, and Cas13a to test if our system would work in vivo . Additionally, we wanted to have RFP as the fluorescent protein to serve as a specificity control instead of transforming the plasmid containing GFP. We were unsuccessful in getting all three plasmids to transform, but did succeed in getting the fluorescent proteins and crRNA containing plasmids to transform together.
As an alternative experiment, our team grew cells that expressed the Lbu and Lwa Cas13a protein overnight. We also grew cells that expressed dual plasmids; GFP and crRNA Lwa; GFP and crRNA Lbu; RFP and crRNA Lwa; and RFP and crRNA Lbu. We then lysed the cells that expressed the Cas13a proteins using a French Press and clarified the lysate via centrifugation. Following this, our team pipetted in a 1:1 ratio of clarified cell lysate: fluorescent protein and crRNA into a 96 well plate. This allowed us to observe if there would be an effect from the CRISPR Cas13a system on the fluorescent proteins. We observed that in our optical density data, both dual plasmid systems for GFP and RFP had stunted growth in comparison to only E. coli cells expressing GFP or RFP or no plasmid (Figure 5C). Adding the lysate may have caused the death of the culture. We neglected to include replicates of the dual plasmid system without adding lysate to observe how that grew. This would be beneficial for any future experiments. Alternatively, there may have been some effect of the protein in the lysate on the GFP fluorescence (Figure 5B). However, we are unsure of the specificity due to the potential of the RFP not folding correctly in vivo as demonstrated by the substantial standard deviation seen in our replicates (Figure 5A).
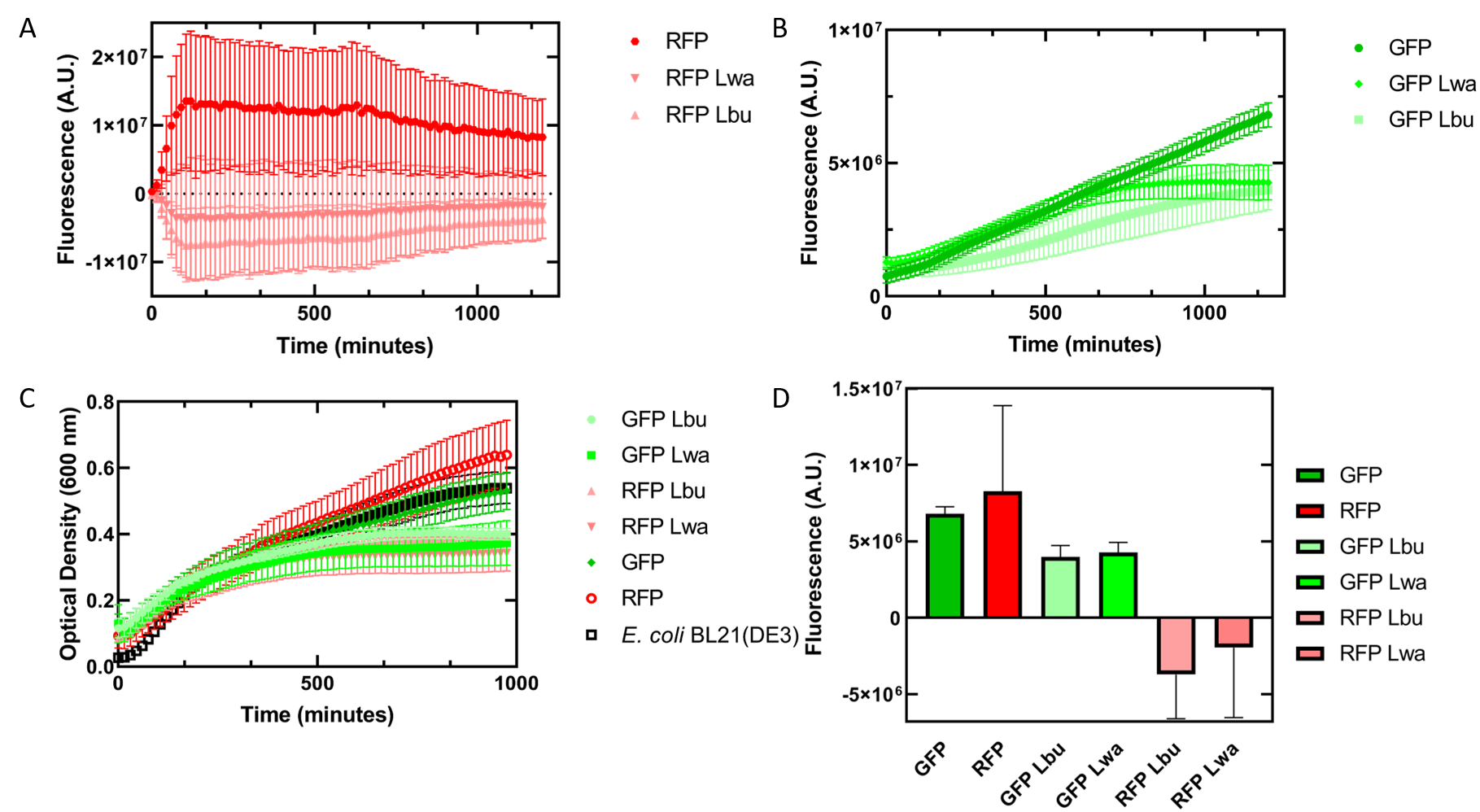 Figure 6. In vivo fluorescence assay of E. coli BL21(DE3) cells containing fluorescent protein and crRNA plasmids and E. coli Rosetta(DE3) cell lysate of overexpressed Cas13a proteins. This assay was conducted with 3 biological replicates and 3 technical replicates. (A) Fluorescence of E. coli cells containing only an RFP expressing plasmid, or dual plasmid expression of RFP and crRNAs from Lwa and Lbu that target GFP and the respective cell lysate containing the appropriate Cas13a. RFP excitation was at 558 nm and emission at 583 nm. (B) Fluorescence of E. coli cells containing only a GFP expressing plasmid, or dual plasmid expression of GFP and crRNAs from Lwa and Lbu that target GFP and the respective cell lysate containing the appropriate Cas13a. GFP excitation was at 475 nm and emission at 508 nm. (C) Optical density of E. coli cells expressing GFP, RFP, dual plasmid systems mentioned previously, or only E. coli BL21(DE3) cells with absorbance measured at 600 nm. (D) Relative fluorescence at maximum excitation at 81 minutes. GFP excitation was at 475 nm and emission at 508 nm and RFP excitation was at 558 nm and emission at 583 nm.
Figure 6. In vivo fluorescence assay of E. coli BL21(DE3) cells containing fluorescent protein and crRNA plasmids and E. coli Rosetta(DE3) cell lysate of overexpressed Cas13a proteins. This assay was conducted with 3 biological replicates and 3 technical replicates. (A) Fluorescence of E. coli cells containing only an RFP expressing plasmid, or dual plasmid expression of RFP and crRNAs from Lwa and Lbu that target GFP and the respective cell lysate containing the appropriate Cas13a. RFP excitation was at 558 nm and emission at 583 nm. (B) Fluorescence of E. coli cells containing only a GFP expressing plasmid, or dual plasmid expression of GFP and crRNAs from Lwa and Lbu that target GFP and the respective cell lysate containing the appropriate Cas13a. GFP excitation was at 475 nm and emission at 508 nm. (C) Optical density of E. coli cells expressing GFP, RFP, dual plasmid systems mentioned previously, or only E. coli BL21(DE3) cells with absorbance measured at 600 nm. (D) Relative fluorescence at maximum excitation at 81 minutes. GFP excitation was at 475 nm and emission at 508 nm and RFP excitation was at 558 nm and emission at 583 nm.
User Reviews
UNIQac8684d7ffa4ff44-partinfo-00000000-QINU UNIQac8684d7ffa4ff44-partinfo-00000001-QINU
