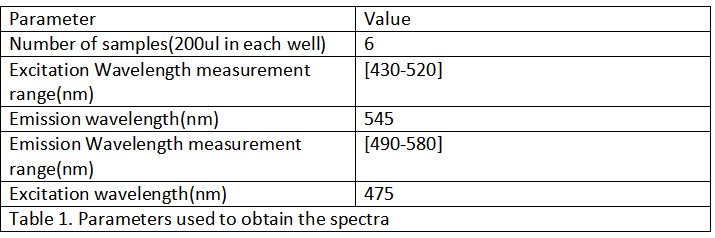
Part:BBa_K1819006
Constitutive promoter (J23106) and GFP (S05308)
Constitutive promotor from Anderson library (BBa_J23106) assembled to green fluorescent protein attached to linker (BBa_S05308). This part was used to characterize BBa_K1819000 and compare its efficiency to green fluorescent protein derived from jellyfish Aequeora victoria wild-type GFP (BBa_E0040).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 717
Characterization by 2019 team XJTU-CHINA
XJTU-CHINA 2019 employed pJ23106 as a constitutive promoter of a photosensitive protein -- Cph8 (BBa_K2598006) which is the key part of our light-control system, so we plan to quantitatively characterize this promoter BBa_J23106 in order to acquire parameters in realistic conditions which can be used in modelling. We use part BBa_K1819006, which is consist of GFP (BBa_K1819000) and its promoter J23106. We have experimentally validated this part in three different chassis of E. coli : DH5α, S17-7 and BL21. We also conduct the scanning of excitation wavelength and emission wavelength at different of time point, trying to obtain the precise emission and excitation spectra in the conditions of our equipment, and evaluate the possible influence of hosts on its spectra. This validation will facilitate the measurements of this GFP with different regulatory parts we assembled. The protocol for bacteria cultivation and fluorescent intensity test parameters are listed below, and Figure 1~Figure 4 were achieved.
Day1: Transform pSB1K3-K1819006 into Escherichia coli DH5α, S17-7 and BL21 respectively. Day2: Inoculate the transformants in 5-10 mL LB medium with chloramphenicol (25 ug/ml) which was cultured for 12h. Day3: 1. Inoculate the recombinant 1% of E.coli into fresh LB medium with chloramphenicol, and culture at 37℃,200 rpm 2. Measure OD600 and GFP fluorescent intensity (RFU) of the bacteria every 2 hours using 200 ul of the culture broth with six parallel groups. 3. We will test (1) OD600, (2) GFP emission scan curve from 495 nm to 580 nm when the excitation wavelength was fixed at 475 nm, (3) GFP excitation scan curve from 430 nm to 520 nm with the fixed emission at 545 nm.
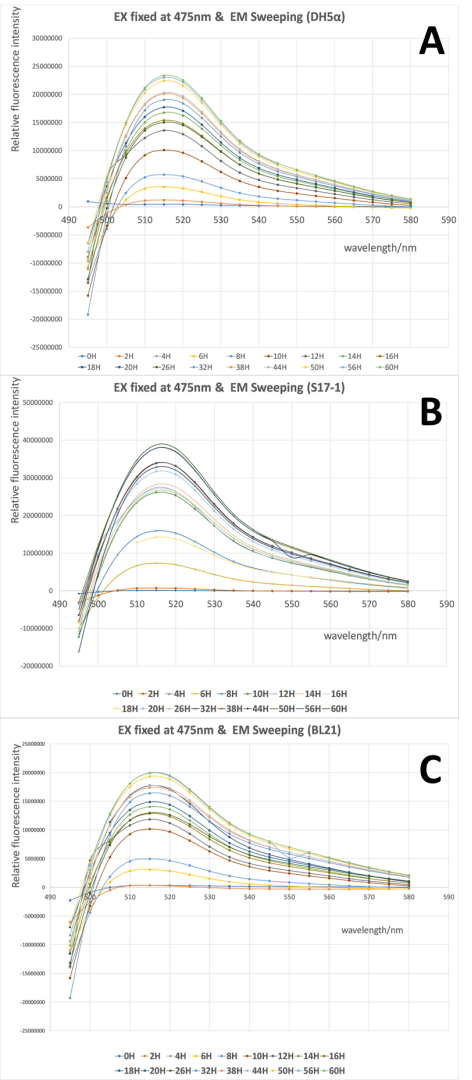
From Fig 1 it is clear to show that the intensity of GFP was gradually increased during the 60h cultivation in all three strains, and the wavelength with highest emission located around 515 nm when the excitation wavelength was fixed at 475 nm. There was no significant difference in the spectra profile of GFP between three E. coli strains, while the intensity of GFP varied in three strains, with the highest intensity in S17-1 at 3.9x107, and lowest in BL21 at 2.0x107.

When the scanning of excitation wavelength was performed with emission wavelength was fixed at 545 nm, similar results were achieved. The intensity of GFP was stably increased and the wavelength with highest excitation was around 505 nm when the emission wavelength was fixed in all three stains. And the three strains shared quite similar spectra profile of GFP, but the intensity of GFP differed, with the highest intensity in S17-1 at 2.6x107, and lowest in BL21 at 1.3x107.
All in all, our validation of the emission and excitation spectra of this GFP in three different recombinant E.coli, provides us the precise wavelength for testing of our parts, as well as demonstrated that the host will not affect the spectra profile of GFP but influence the intensity. From the results above, we can could that the maximum excitation wavelength of the GFP in this part is around 505nm, and the maximum emission wavelength is around 515 nm.
Additionally, we also measured the fluorescent curve using the max ex and max em we obtained above (ex=505 nm, em=515 nm). The results are as follows:
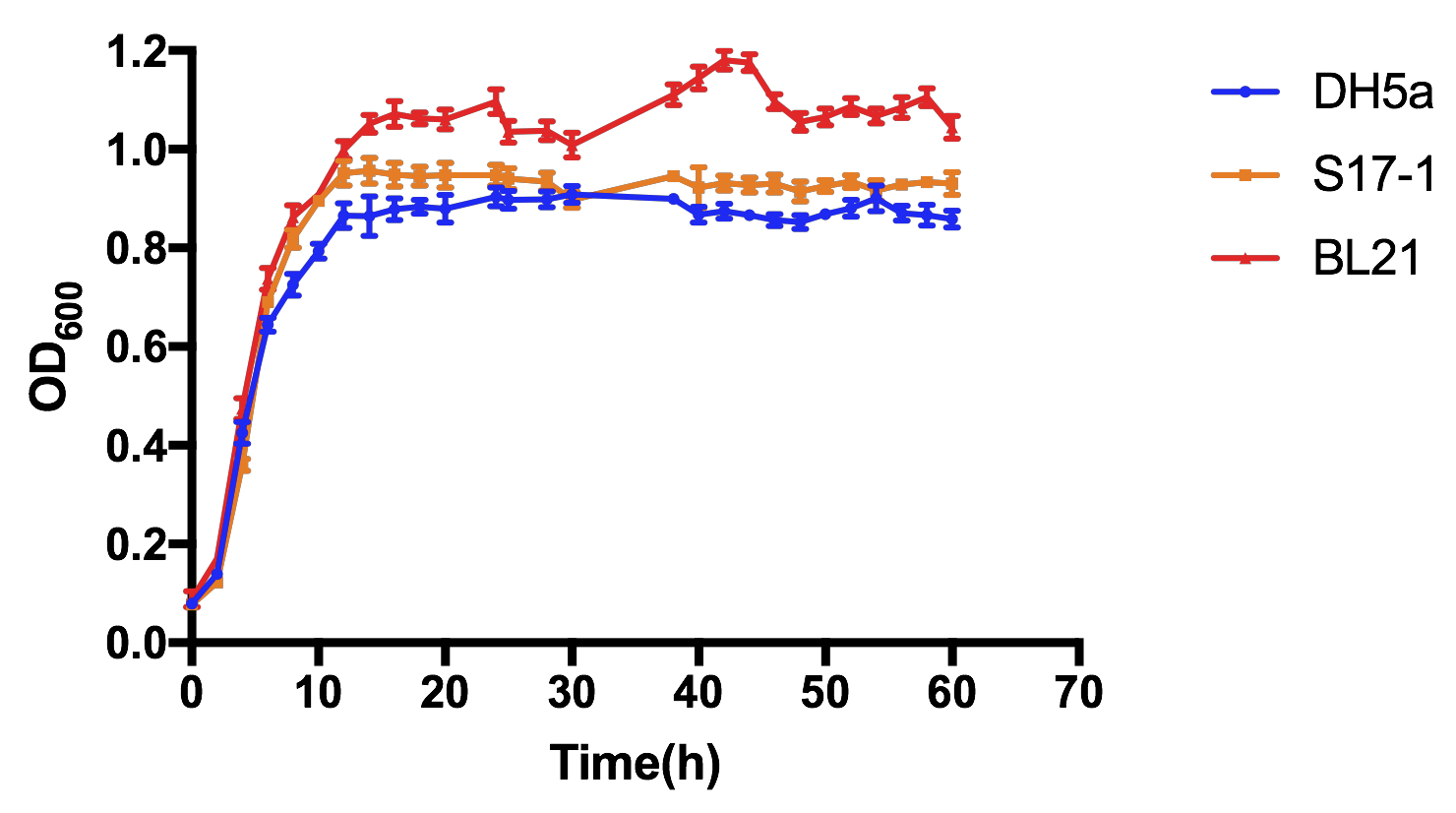
As shown in Fig 3, all the three E. coli experienced the basic cell growth of very short lag phase, fast log phase and quite long stationary phase during 60-h cultivation. OD600 of all three strains basically reached stability after about 14h cultivation, and it is clearly showed that BL21 had the most vigorous growth, S17-1 the second, while DH5α had the slowest growth, but there was no significant difference between these three, in case of lab-scale cultivation.
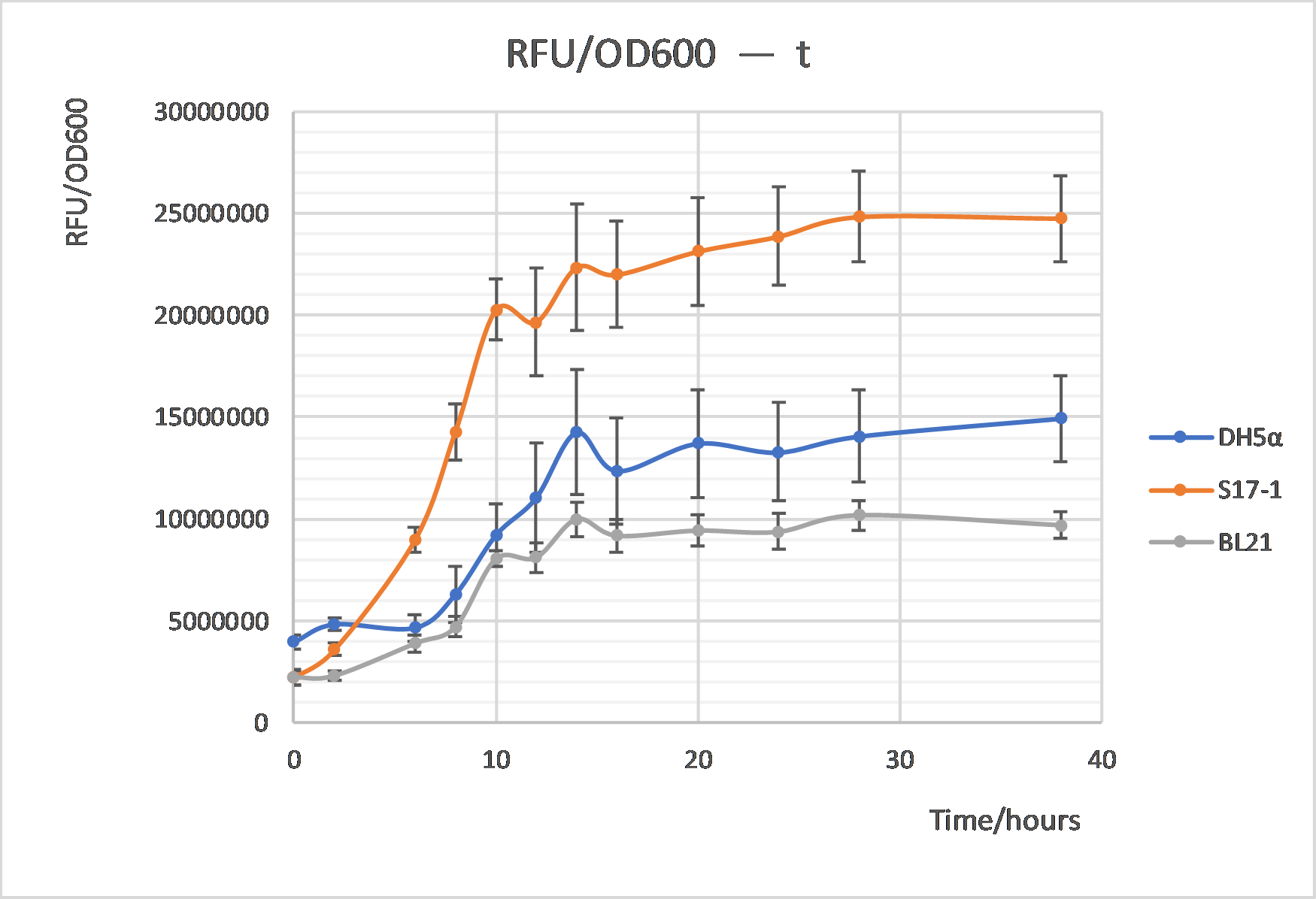
As shown in Fig 4, the GFP intensity changes per cell were also be compared between these three strains, which shared the similar trends with cell growth. The GFP intensity per cell turned to be stable after about 12h cultivation. And recombinant S17-1 exhibited highest intensity among three strains, while BL21 is the least, indicating S17-1 somehow could be a better host for the measurement of this GFP-harboring parts based on our results.
//function/reporter
//function/reporter/fluorescence
| color | Green |
| direction | Forward |
| emission | |
| emit | 511 |
| excitation | |
| excite | 501 |
| kegg | |
| lum | |
| protein | GFPmut3b |
| swisspro |