Part:BBa_K1789023
IAA(+/+)S3
This is a device which contains the sequence of the recombination of TALE1 and IAAM, the recombination of TALE2 and IAAH and scaffold 3. Through this device, we can test and verify that our scaffold can strengthen the function of IAA.
Usage and Biology
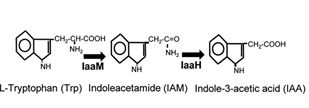
Indole-3-acetic acid (IAA), also known as auxin, is a plant hormone, which can promote growth and protect the soil from erosion. IAA can be synthesized through IAM pathway, originated from Pseudomonas savastanoi. Two important enzymes are contained in this pathway, AKA IaaM and IaaH.
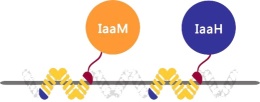
In this device, the TALE1-fused IAAM and the TALE2-fused IAAH are incorporated in the E.coli due to the specific binding of TALE proteins and scaffold3, which could efficiently guide the enzymes around the DNA scaffolds, to enrich the local enzyme concentration, and promote the rate of reaction. As a supplement, distance between IAAM and IAAH in this device is more than that in IAA(+/+)S2 (the part BBa_K1789022 ).
Sequence and Features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2724
Illegal BamHI site found at 2311
Illegal BamHI site found at 3669
Illegal BamHI site found at 6072 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 2431
Illegal NgoMIV site found at 7088
Illegal AgeI site found at 7571
Illegal AgeI site found at 7633
Illegal AgeI site found at 7695
Illegal AgeI site found at 7757
Illegal AgeI site found at 7819
Illegal AgeI site found at 7881
Illegal AgeI site found at 7943 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1304
Illegal BsaI.rc site found at 5166
Illegal BsaI.rc site found at 5370
Experimental Validation
Sequencing
This part is sequenced as correct after construction.
RT-PCR Analysis
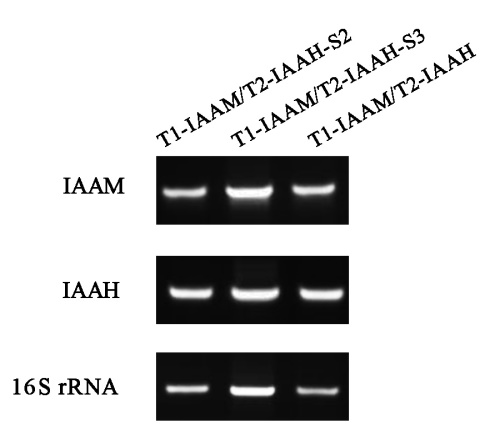
To evaluate whether the expression variation of IAAM and IAAH can be promoted by the specific binding of TALE proteins and scaffolds, the RT-PCR analysis was conducted. RT-PCR analysis showed that the increase of IAA production in scaffold system was not owing to the expression variation of IAAM and IAAH. (Fig. 1)
Fig. 1 RT-PCR analysis for determination of IAAM and IAAH expression in different TALE-IAA-scaffold groups. The cDNA sequence of 16S rRNA was amplified as standard.
Sandwich ELISA Analysis
To evaluate whether the rate of reaction can be promoted by the specific binding of TALE proteins and scaffolds, the sandwich ELISA analysis was conducted.
The TALE1-IAAM/TALE2-IAAH-Scaffold2 and TALE1-IAAM/TALE2-IAAH-Scaffold3 plasmids were transformed to E.coli BL21 (DE3), cultured and then IPTG induced overnight. The TALE1-IAAM/TALE2-IAAH transformed group was invoked as no scaffold control.
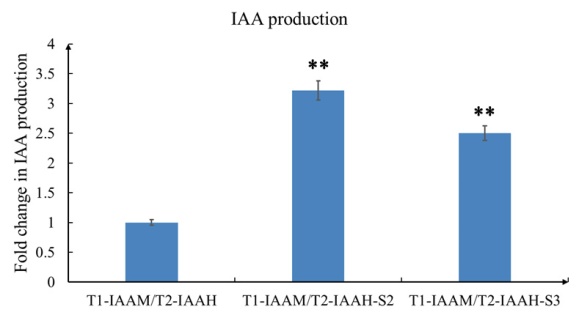
By sandwich ELISA analysis, both of the scaffold harboring groups showed significantly higher production of IAA than the no scaffold control group (>3.2 fold in TALE1-IAAM/TALE2-IAAH-Scaffold2 and >2.5 fold in TALE1-IAAM/TALE2-IAAH-Scaffold3, p<0.01, Fig.2).
Fig. 2 The production of IAA was determined by sandwich ELISA after overnight culture of E.coli with IPTG induction. After TMB coloring and reaction termination, the color was measured spectrophotometrically at a wavelength of 450 nm. The concentration of IAA in the samples is then determined by comparing their OD450 to the standard curves. The relative IAA concentration of TALE1-IAAM/TALE2-IAAH no scaffold control group was set arbitrarily at 1.0, and the levels of other groups were adjusted correspondingly. All of the data represent results obtained from at least three independent experiments. **p<0.01.
Summary
According to these results, it could be demonstrated that the TALE-DNA scaffold system can effectively increase the IAA production through an IAAM-IAAH metabolic pathway. Thus, TALE-DNA scaffold system might be an efficient accelerator to promote the rate of heterologous metabolic pathway in prokaryotic chassis.
| None |