Part:BBa_K1789003
GFP1(with termination codon)
This part is the Amino Half of GFP with termination codon
Usage and Biology
Bimolecular fluorescence complementation (BiFC) means two non-fluorescent complementary fragments of the fluorescent protein can reassemble to form a fluorescent complex and restore fluorescence when they are fused to two proteins that interact with each other.
BiFC analysis has been used to study interactions among a wide range of proteins in many cell types. The study of interactions and post-translational modification of the protein makes people master the biological regulatory mechanism more. Interactions in protein are also highly valued, so there have been a number of related technologies having different characteristics and applications[1-2].
Such as GFP, after the sequence of certain sites in interchanges with the amino-terminal or carboxy-terminal sequence loop, it could still be able to fold correctly to form the structure of the chromophore and maintain fluorescence properties[3-5]
In our project, we plan to fuse the GFP1 to the C-terminal of TALE1 protein. However, the sample of BBa_ I715019 provided by the 2015 DNA distribution does not have a termination codon on its 3’ terminal to stop the translation. To fix this, we designed a pair of primers to add a termination codon on the 3’ terminal of BBa_ I715019 to further extend its usage.
Sequence and Features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Experimental Validation
This part is validated through four ways: enzyme cutting, PCR, Sequence, and functional testing
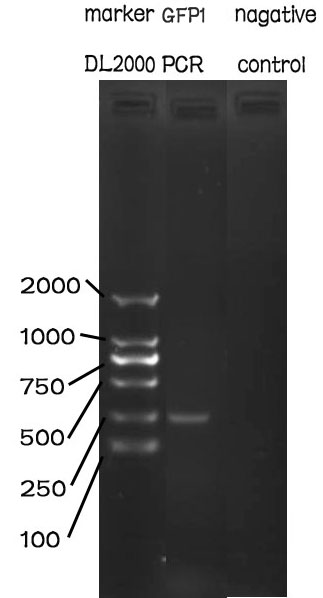
PCR
Methods
The PCR is performed with Premix EX Taq by Takara.
F-Prime: 5’- GAATTCGCGGCCGCTTCTAGAATGC-3’
R-Prime: 5’- GGACTAGTATTATTGTTTGTCTGCC-3’
The PCR protocol is selected based on the Users Manuel. The Electrophoresis was performed on a 1% Agarose glu.
The result of the agarose electrophoresis was shown on the picture above.
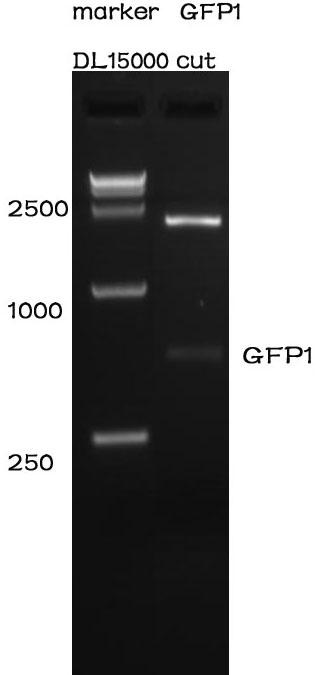
Enzyme cutting
Methods
After the assembly ,the plasmid was transferred into the Competent E. coli DH5α). After culturing overnight in LB,we minipreped the plasmid for cutting. The preparation of the plasmid was performed with TIANprep Mini Plasmid Kit from TIANGEN. The cutting procedure was performed with EcoRI and XbaI restriction endonuclease bought from TAKARA.
The plasmid was cutted in a 20μL system at 37 ℃ for 2 hours. The Electrophoresis was performed on a 1% Agarose glu.
Results
The result of the agarose electrophoresis was shown on the picture above.
Functional Test
This part is tested together with the part BBa_K1789004, in the device BBa_K1789019.
The result shows that this part can shine with GFP2.
References
[1] Drewes G, Bouwmeester T.Global approaches to protein – protein interaction[ J ]. Curr Opin Cell Biol, 2003, 15(2): 199-205.
[2] Collura V, Boissy G. From protein -protein complexes to interactomics [ J ]. Subcell Biochem, 2007, 43: 135-83.
[3] M isteli T, Spector DL. Application of the green fluorescent protein in cell biology and biotechnology[ J ]. Nat Biotechnol, 1997, 15( 10): 961 - 964.
[4] Baird G. S, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins[ J ]. Proc Natl Acad Sci USA , 1999, 96( 20): 11241-11246.
[5] Ghosh I, Hamilton AD, Regan L. Antiparallel leucinezipper –directed protein reassembly: application to the green fluorescent protein[ J ]. J Am Chem Soc, 2000, 122: 5658-5659.
//function/reporter/fluorescence
| color | Green |