Part:BBa_K1701006
CotC
CotC, one of the protein components of the spore coat of Bacillus subtilis, with other proteins like Cot B, Cot F, can be used as a fusion partner to express proteins or polypeptides on the spore surface.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Background
The Gram positive bacterium Bacillus subtilis has been extensively studied as a model prokaryotic system with which to understand gene regulation and the transcriptional control of unicellular differentiation. This organism is regarded as a non-pathogen and is classified as a novel food which is currently being used as a probiotic for both human and animal consumption.
B. subtilis can divide symmetrically to make two daughter cells (binary fission), or asymmetrically, producing a single endospore that can remain viable for decades and is resistant to unfavourable environmental conditions such as drought, salinity, extreme pH, radiation, and solvents. The endospore is formed at times of nutritional stress, allowing the organism to persist in the environment until conditions become favourable. Prior to the process of sporulation the cells might become motile by producing flagella, take up DNA from the environment, or produce antibiotics. These responses are viewed as attempts to seek out nutrients by seeking a more favourable environment, enabling the cell to make use of new beneficial genetic material or simply by killing of competition.
Under stressful conditions, such as nutrient deprivation, B. subtilis undergoes the process of sporulation to ensure the survival of the species. This process has been very well studied and has served as a model organism for studying sporulation.
Biology
The distinguishing feature of Bacillus subtilis is that it produces an endospore as part of its developmental life cycle when starved of nutrients.
The mature spore, when released from its mother cell can survive in a metabolically dormant form indefinitely. The spore offers unique resistance properties and can survive extremes of temperature, dessication and exposure to solvents and other noxious chemicals.
The protein component of the spore coat, such as Cot B, Cot C, Cot F, can be used as a fusion partner to express proteins or polypeptides on the spore surface.
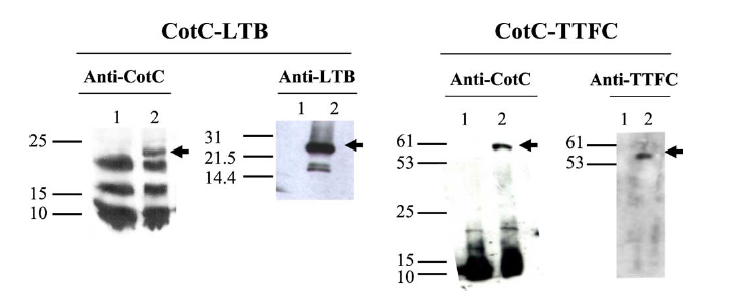
Figure 1.Western blot of proteins extracted from purified spores of strains IM201 (CotC-LTB) and RH114 (CotC-TTFC) and reacted against anti-CotC, anti-LTB and anti-TTFC antibodies. Fifteen micrograms of total proteins from strains PY79 (lanes 1 in all panels), IM201 (lanes 2, Panel CotC-LTB) and RH114 (lanes 2 Panel CotC-TTFC) were fractionated on 10% polyacrylamide gel and upon electro-transfer on nitrocellulose membranes, reacted with primary anti-rabbit antibodies, then with secondary antibodies and visualised as described in Section 2. Arrows point to fusion proteins. Molecular weight markers (kDa) are indicated.
Part uses
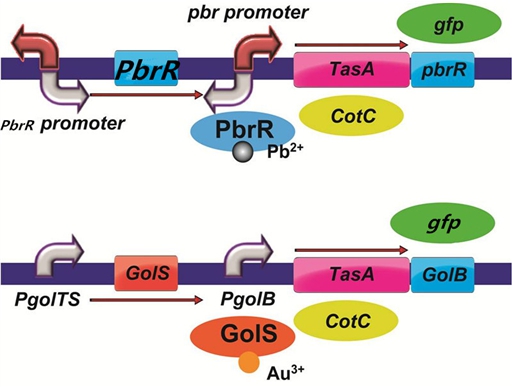
In order to improve the efficiency of heavy metal adsorption that our project aims at, we decided to use the cell surface-display strategy by using CotC, the major protein of the spore component in B. subtilis as fusion partner to display the heavy metal binding proteins(GolB, PbrR and SUP).
When conditions get adverse for the growth of cells, B. subtilis produce spores to survive. If we express the metal binding proteins along with the coat protein CotC, spores will be capable of heavy metal adsorption.
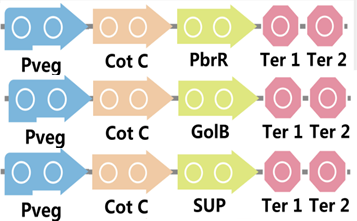
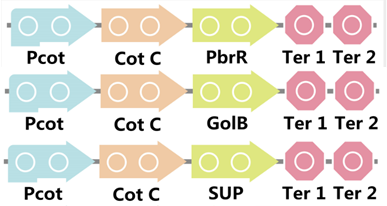
To realize this goal, we have designed three pathways shown in Figure 2, which are used in our project. In order to improve the efficiency of the expression of the three pathways, we utilized a constitutive promoter Pveg, a biobrick part in the biological parts of iGEM to replace the promoter Pcot, which is involved in the three pathways in Figure 3.
Figure 2. Three pathways designed by our group involving CotC part.
Figure 3. Three pathways designed by our group involving CotC part but replacing Pcot with Pveg.
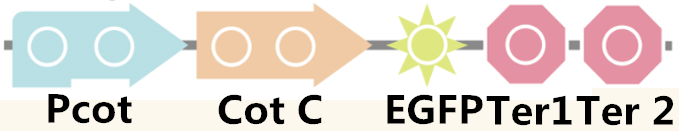
We also examined the ability of the expression by using EGFP, the fluorescence protein, and the pathway is shown in Figure 4.
Figure 4. The pathway used for fluorescence measurement.
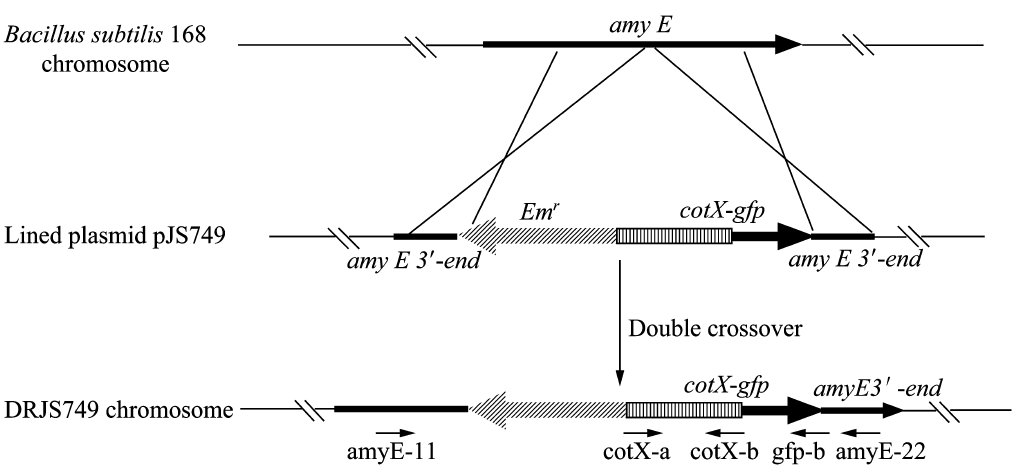
We will construct our plasmid in E.coli first and affirm the feasibility of the pathways, after which we will transform the plasmid into B.subtilis. After linearization of the plasmid, we can integrate the target genes into the chromosomes of B.subtilis utilizing homologous recombination. The process and the shuttle plasmid of our project can be shown as follows.
Figure 5. The process to integrate the target genes into the chromosomes of B.subtilis utilizing homologous recombination.
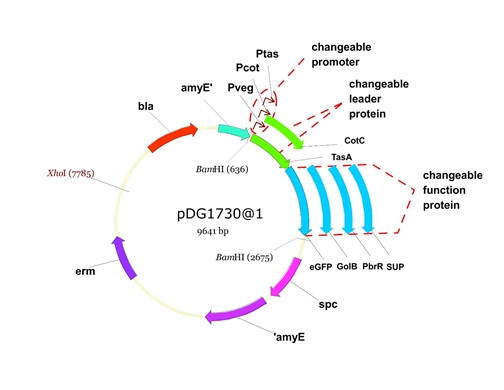
Figure 6. The shuttle plasmid designed by our project including the preceding pathways.
To improve the selectivity of heavy metal(such as lead and gold) adsorption, we had a plan to design two pathways as follows. With the heavy metal specific promoter, the heavy metal-binding spore protein including CotC can be expressed only when there is specific heavy metal in the solution. In addition, we had a plan to do a pre-test to demonstrate the feasibility of the pathways by replacing the heavy metal binding protein with the reporter protein GFP.
Figure 7. The designed pathways to improve the selectivity of lead and gold adsorption.
Results
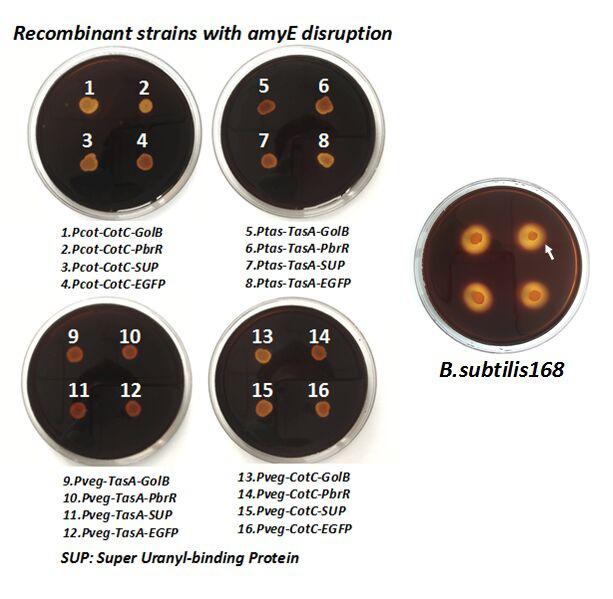
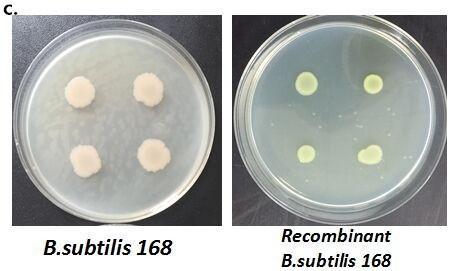
After we constructed the 16 kits in E.coli, we inserted them into the shuttle plasmid pDG1730 which can transfer from E.coli to B. subtilis. We insert our fusion genes between two sequences of the amylase genes on the plasmid. By homologous recombination, our fusion genes will replace the amylase gene on the genome of B. subtilis. In order to test and verify whether homologous recombination is successful, we built the following three steps. Antibiotic selection, bacterial PCR analysis and amylase activity analysis. After a series of test and verification experiments, we have successfully inserted the 16 kits into the genome of B. subtilis. The following figure shows the result of the amylase activity analysis. The picture on the right is the control group while the pictures on the left are the experiment groups. When we applied iodine on the plate with starch, there would be hydrolysis cycles on the plate because the α-amylase gene wasn’t replaced. However, there wouldn’t be hydrolysis cycles on the plate if homologous recombination was successful.
Figure 8. Result of homologous recombination.
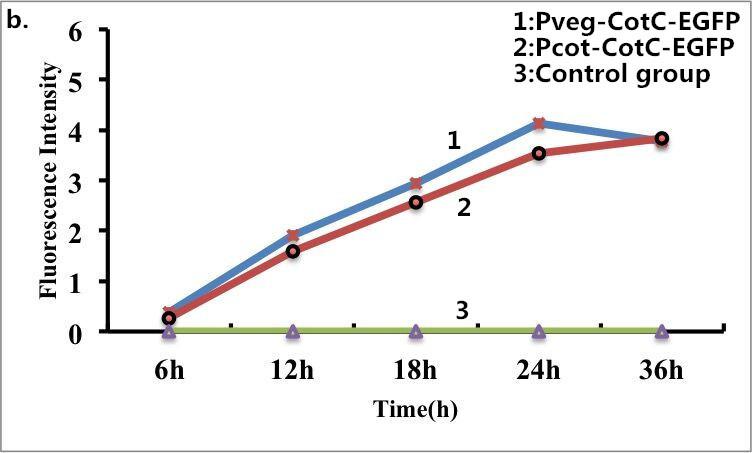
B. subtilis cells carrying fusion proteins (CotC-EGFP) were fixed for visualization under a fluorescence microscope. The same batch of B. subtilis cells without fusion proteins was used as s control. Strong fluorescence was observed when the B. subtilis cells expressed the fusion proteins (CotC-EGFP), whereas no fluorescence was detected for the control group. Taken together, these results confirmed the fusion proteins can express well on the surface of B. subtilis. The following pictures show the results of fluorescence measurements.
Figure 9. Fluorescence measurement of B. subtilis cells containing the fusion proteins. b) Fluorescence intensity of the fusion proteins Pveg-CotC-EGFP and Pcot-CotC-EGFP. c) A visible photograph of b).
References
[1] E.M.F. Mauriello et al. / Vaccine 22 (2004) 1177–1187Mauriello EMF, Duc LH, Isticato R, Cangiano G, Hong HA, De Felice M, Ricca E, Cutting SM: Display of heterologous antigens on theBacillus subtilisspore coat using cotc as a fusion partner.Vaccine 2004, 22:1177–1187.
| biology | Bacillus subtilis |