Part:BBa_K1701004
SUP
A uranyl-binding protein (SUP) with potential uranyl binding sites which are selective for uranyl.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Background
Uranyl (UO22+), the predominant aerobic form of uranium, is present in the ocean at a concentration of ∼3.2 parts per 109 ; however, the successful enrichment of uranyl from this vast resource has been limited by the high concentrations of metal ions of similar size and charge, which makes it difficult to design a binding motif that is selective for uranyl. It has been reported that the design and rational development of a uranyl-binding protein (SUP) using a computational screening process in the initial search for potential uranyl binding sites.
Protein sturcture
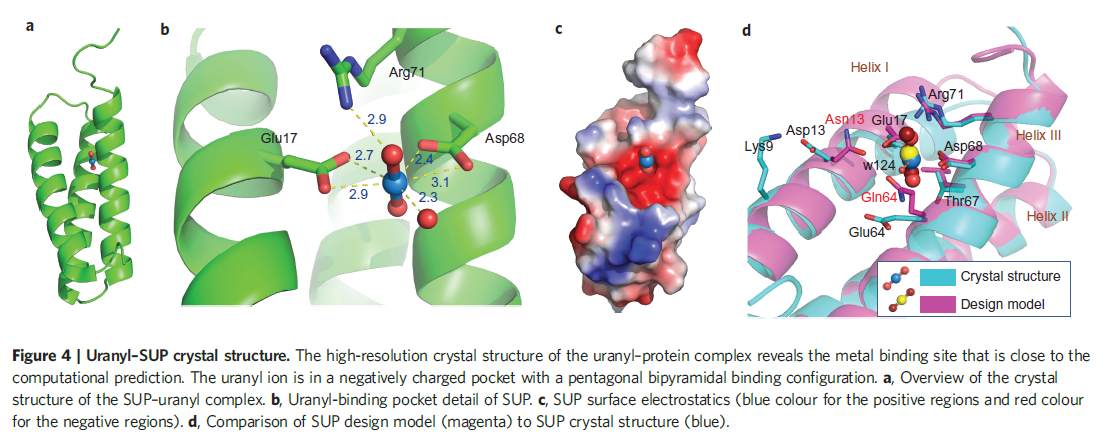
The picture below shows the crystal structure of the designed uranyl-binding protein SUP. See more details about the crystal stucture of SUP in the paper we mention in the Reference.
Figure 1. Uranyl-SUP crystal stucture.
Protein Charecteristics
The engineered protein is thermally stable and offers very high affinity and selectivity for uranyl with a Kd of 7.4 femtomolar (fM) and >10,000-fold selectivity over other metal ions. Scientists also demonstrated that the uranyl-binding protein can repeatedly sequester 30–60% of the uranyl in synthetic sea water.
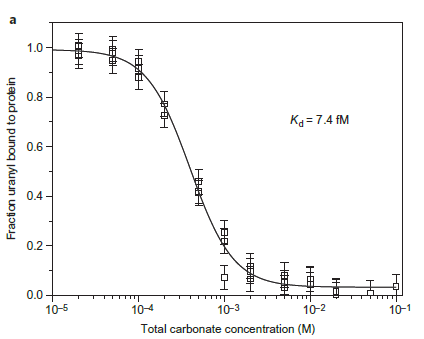
Figure 2(a). Competition assay of SUP versus total carbonate for uranyl yields a Kd of 7.4 fM at pH 8.9. The final solution of each point contained 10 mM protein, 10mM UO2 2+ and different concentrations of carbonate.
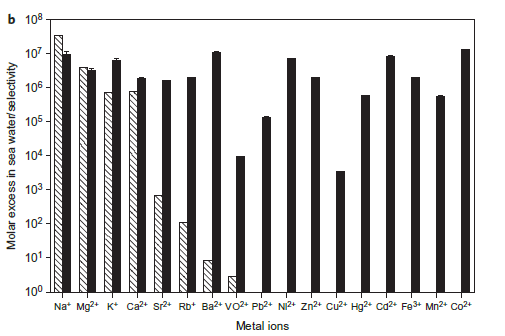
Figure 2(b).Binding selectivity of SUP for uranyl over various other metals relevant to sea water extraction. Hatched columns, molar excess of ions in sea water; filled columns, selectivity of metal ions to uranyl by competition assay.
Part uses
we heterologously expressed the Uranyl-binding chaperon SUP gene into Bacillus subtilis. Hence, we displayed SUP on Bacillus subtilis biofilm or its spore surface to construct a whole-cell absorption specific for Uranyl, which is able to be easily recovered and therefore is good for application outside of laboratory settings. What’s more, it can adsorb Uranyl under both benign conditions and adverse circumstances.
We used TasA, the major protein of the biofilm matrix component in B. subtilis, and CotC, the B.subtilis spore component, as fusion partner to display SUP.
When nutrition is sufficient, we can use plant polysaccharides to induce the formation of biofilm, on which protein TasA can be simultaneously expressed with SUP. Besides, when conditions get adverse for the growth of cells, B. subtilis produce spores to survive. If we express the SUP along with the coat protein CotC, spores will be capable of Uranyl adsorption. Therefore, B. subtilis can adsorb Uranyl under both benign conditions and adverse circumstances, which tremendously improve the efficiency of Uranyl adsorption.
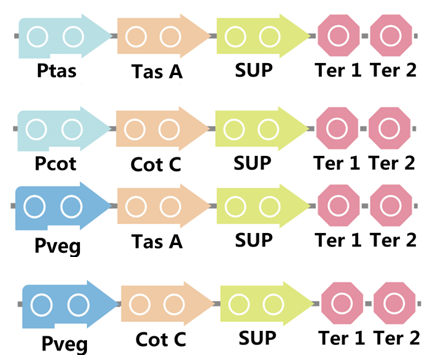
To realize this goal, we have designed four pathways shown in Figure 2, which are used in our project. In order to improve the efficiency of the expression of the two former pathways, we utilized a constitutive promoter Pveg, a biobrick part in the biological parts of iGEM used by us to replace the promoter Ptas and Pcot, which is involved in the two latter payways.
Figure 3. Four pathways designed by our group involving SUP part.
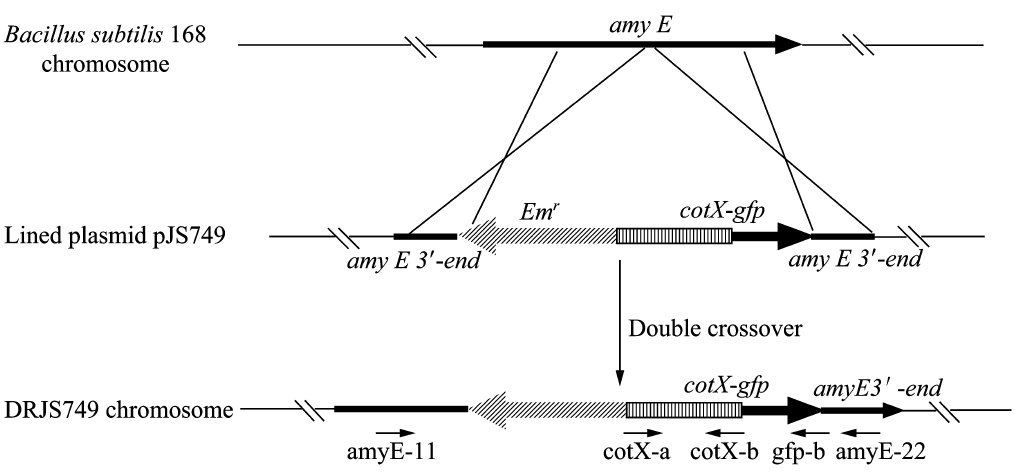
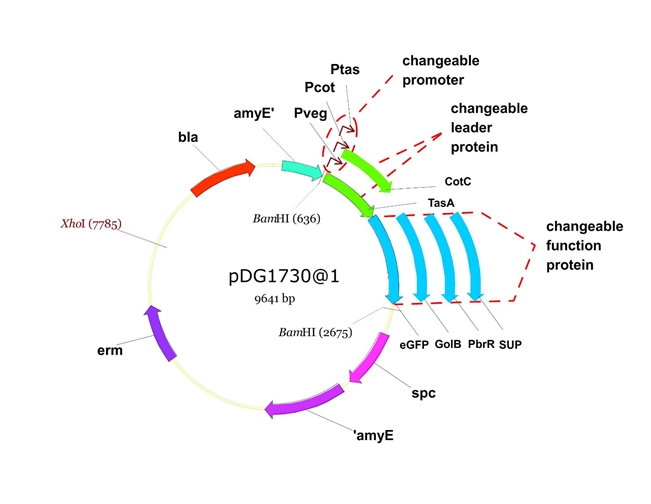
We will construct our plasmid in E.coli first and affirm the feasibility of the pathways, after which we will transform the plasmid into B.subtilis. After linearization of the plasmid, we can integrate the target genes into the chromosomes of B.subtilis utilizing homologous recombination. The process and the shuttle plasmid of our project can be shown as follows.
Figure 4. The process to integrate the target genes into the chromosomes of B.subtilis utilizing homologous recombination.
Figure 5. The shuttle plasmid designed by our project including the preceding pathways.
Results
Homologous recombination
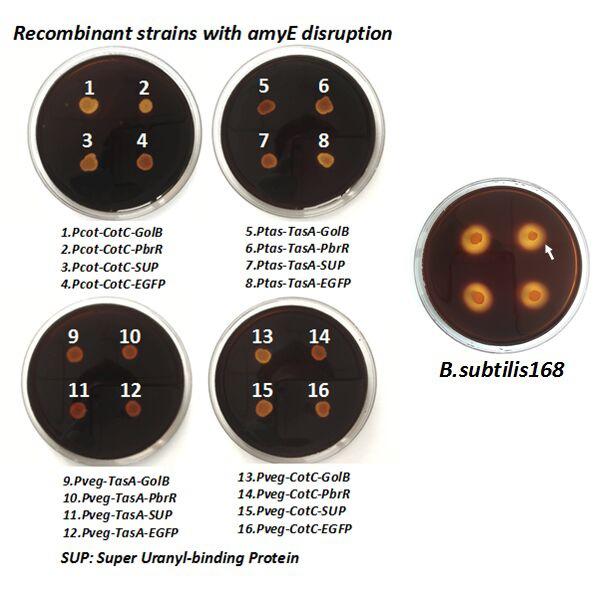
After we constructed the 16 kits in E.coli, we inserted them into the shuttle plasmid pDG1730 which can transfer from E.coli to B. subtilis. We insert our fusion genes between two sequences of the amylase genes on the plasmid. By homologous recombination, our fusion genes will replace the amylase gene on the genome of B. subtilis. In order to test and verify whether homologous recombination is successful, we built the following three steps. Antibiotic selection, bacterial PCR analysis and amylase activity analysis. After a series of test and verification experiments, we have successfully inserted the 16 kits into the genome of B. subtilis. The following figure shows the result of the amylase activity analysis. The picture on the right is the control group while the pictures on the left are the experiment groups. When we applied iodine on the plate with starch, there would be hydrolysis cycles on the plate because the α-amylase gene wasn’t replaced. However, there wouldn’t be hydrolysis cycles on the plate if homologous recombination was successful.
Figure 6. Result of homologous recombination.
Bio-adsorption
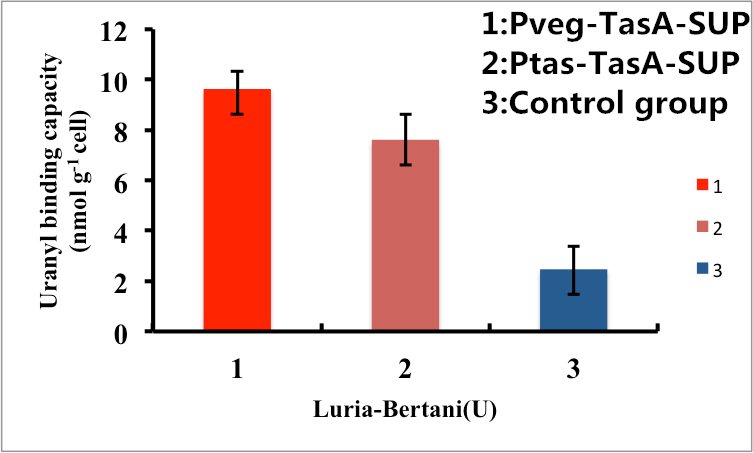
As shown in Fig. 7, B. subtilis bacteria with the TasA-dependent surface displayed SUP with promoter Pveg were able to adsorb uranyl ions with a capacity of about 8.7 μmol g-1cells, which is 4-fold higher than un-displayed samples. While bacteria with promoter Ptas were able to adsorb uranyl ions with a capacity of about 6.8 μmol g-1cells, which is 3-fold higher than un-displayed samples.
Figure 7. Adsorption of uranyl ions by B. subtilis TasA-dependent surface-displayed SUP protein.
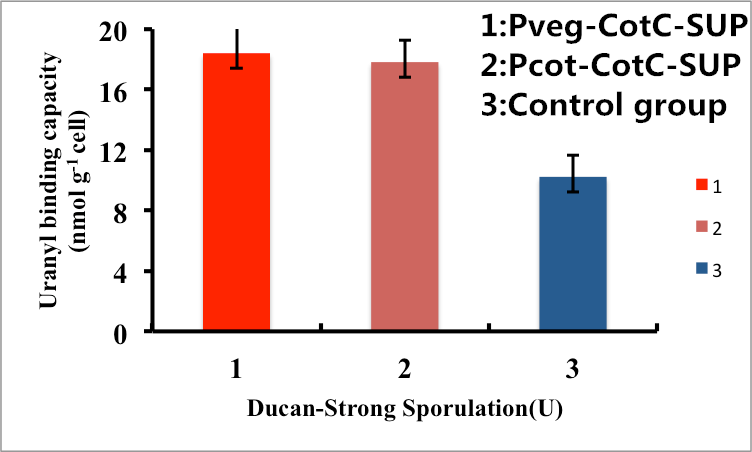
As shown in Fig. 8, B. subtilis endospores with the CotC-dependent surface displayed SUP with promoter Pveg were able to adsorb uranyl ions with a capacity of about 16.5 μmol g-1cells, which is 2-fold higher than un-displayed samples. While bacteria with promoter Pcot were able to adsorb uranyl ions with a capacity of about 16.3 μmol g-1cells, which is 2-fold higher than un-displayed samples.
Figure 8. Adsorption of uranyl ions by B. subtilis endospore CotC dependent surface-displayed SUP protein.
In conclusion, the engineered B. subtilis as well as the endospores worked well in heavy metal adsorption. Besides, it is proved by us that Pveg is a more efficient promoter than Ptas and Pcot, so maybe we can utilize promoter Pveg when engineering B. sublitis.
Improvements
Peking iGEM 2016 has fused this part with triple, quadruple and sextuple spytag, respectively. The fused protein is capable of binding uranyl as well as forming polymer network. When polymer network is formed, the uranyl binding capacity is even evaluated.
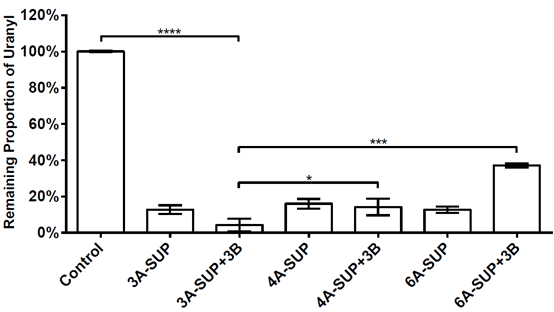
We tested the adsorption capacity of 3A-SUP, 4A-SUP, 6A-SUP and 3A-SUP+3B, 4A-SUP+3B, 6A-SUP+3B in TBS buffer (pH=7.02) against 10μM uranyl. We wanted to do some comparisons and found the best candidate. Cross-linked 3A-SUP+3B can sequester 95.77% of the total uranyl in TBS buffer (pH=7.0, 10μM), showing the best adsorption capacity. Other proteins also can sequester at least 60% of the total uranyl. The standard deviation were calculated from triplicate experiments.
Figure 9. Adsorption capacity of 3A-SUP,4A-SUP,6A-SUP and 3A-SUP+3B,4A-SUP+3B,6A-SUP+3B
If you want to learn more about Peking’s polymer network and the role of SUP in this network, please click here https://parts.igem.org/Part:BBa_K1989000", https://parts.igem.org/Part:BBa_K1989005" or https://parts.igem.org/Part:BBa_K1989006".
References
[1].Zhou, L., et al., A protein engineered to bind uranyl selectively and with femtomolar affinity. Nat Chem, 2014. 6(3): p. 236-41.
[2].Odoh, S.O., et al., UO(2)(2)(+) uptake by proteins: understanding the binding features of the super uranyl binding protein and design of a protein with higher affinity. J Am Chem Soc, 2014. 136(50): p. 17484-94.
| None |