Part:BBa_K150009
ColicinE1 Producer Controlled by 3OC6HSL Receiver Device
The constitutive expressed LuxR protein is able to build a complex with an AHL molecule. [7] This complex functions as transcription factor for the lux pR promoter and activates therefore colicinE1 production. The activation of the lux pR promoter leads additionally to expression of the kil protein, also called lysis protein by readthrough of a terminator located in the colicinE1 operon. Production of the lysis protein leads to lysis of the host cell and colicin release. The colicinE1 immunity protein (imm) located on the opposite strand is expressed under a constitutive promoter and protects the host cell from lysis up to a certain threshold of the lysis protein. (See experience for characterization)
Usage and Biology
Colicin E1 - Mode of Action
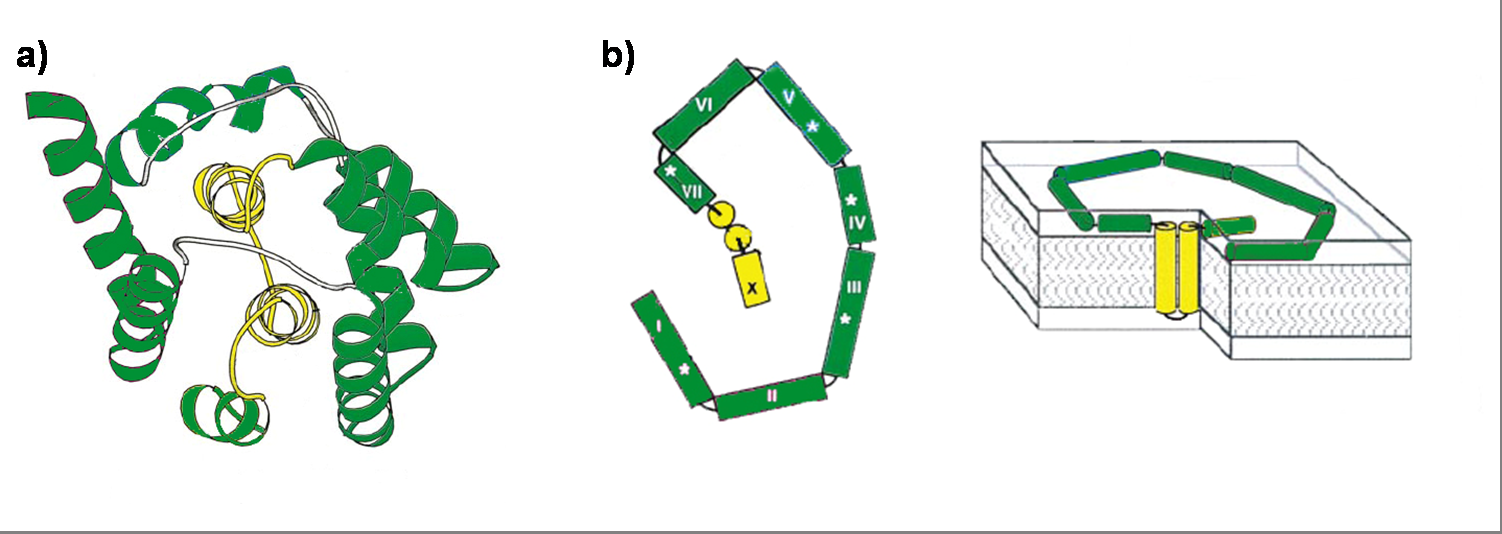
Colicin E1 is a 522 residue containing protein and belongs to the group A colicins. It kills sensitive cells, which harbor no ColE1 plasmid but contain the appropriated receptor BtuB, by forming pores, or more precisely ion channels into the inner membrane. [1] Affected cells can be E. colis or related strains but also some eukaryotic cells have been affected by the toxic activity of colicin. [2] The first step of the lethal action of colicin E1 is binding to the vitamin B12 receptor (BtuB). Colicins are able to kill bacteria by a “one hit” mechanism. This works due to destruction of the target cells cellular energy. The voltage dependant channel in the cytoplasmatic membrane leads to a depolarization of the target cell. The conductance of the channel has been shown to be ~107 ions/channel-sec in 1 M NaCl. [3] Its ability to form pores is given by the C-terminal domain. This domain consists of 10 tightly packed α–helices (see Figure 1). [4], [6] The colicin protein is a water soluble molecule containing mostly amphipathic helices except from two hydrophobic ones. Due to these two hydrophobic helices the protein is able to target the cell membrane and act as a membrane protein.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1083
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1004
Illegal SapI site found at 1389
References
[1] Cramer W. et al. (1990) Structure and dynamics of the colicin E1 channel. Molecular Microbiology 4: 519-526.
[2] Smarda J. et al. (2001) Cytotoxic effects of colicins E1 and E3 on v-myb-transformed chicken monoblasts. Folia Microbiol. (Praha) 47: 11–13.
[3] Bullock J. et al. (1983) Comparison of the macroscopic and single channel conductance properties of colicin E1 and its COOH-terminal tryptic peptide. J. Biol. Chem. 258: 9908–9912.
[4] Elkins P. et al. (1997) A mechanism for toxin insertion into membranes is suggested by the crystal structure of the channel-forming domains of Colicin E1. Structure. 5: 443-458.
[5] Lindeberg M. et al. (2000) Unfolding Pathway of the Colicin E1 Channel Protein on a Membrane Surface. J. Mol. Biol. 295: 679-692
[6] Davies, J. et al. (1975) Genetics of resistance to colicins in Escherichia coli K12: cross-resistance among resistance of group A. J. Bacteriol. 123:102–117
[7] Engebrecht J. et al. (1984) Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 81: 4154–58
| None |
