Part:BBa_K1499503
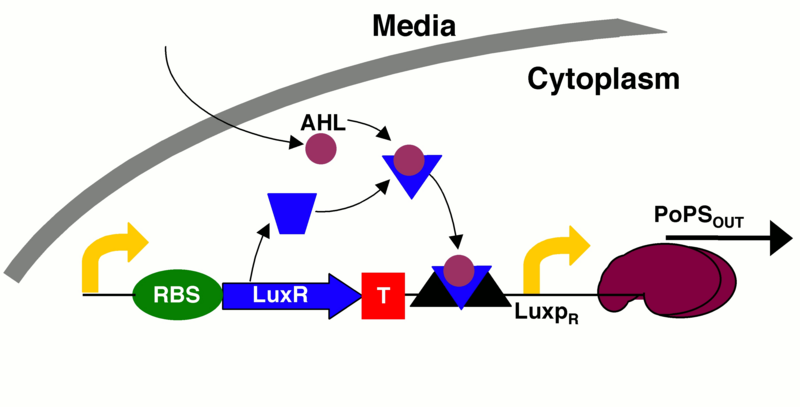
quorum sensing machinery that activates GFP expression with extra terminator
This is the same part as Part:BBa_K1499500, except that it has two more terminators between the luxR gene and the luxPR promoter.
After ligating the BBa_K1499500 construct together and into the BioBrick backbone, we transformed it into the NEB 5-alpha strain of E. coli. The colonies that fluoresced were most likely to have been successfully transformed, and thus these were sent off for sequencing. The sequencing data showed that our construct was correct (see below), so we were able to submit for BioBricking.
Our goal in building this construct is to create a time delay. Since the production of AHL is under the control of a lac-repressible promoter, the cascade should not begin until the promoter is induced. Additionally, once the promoter is activated, time is required for the AHL and luxR molecules to build up and interact with one another. Only after these molecules are present in large enough quantities to interact and bind to the luxPR promoter will GFP be expressed. Thus by replacing GFP with any gene of interest, this construct can be used to create a time delay for the onset of gene expression. For more information on the specifics of the time delay, see the "experience" page.
Usage and Biology
After ligating this construct together and into the BioBrick backbone, we transformed it into the NEB 5-alpha strain of E. coli. The colonies that fluoresced were most likely to have been successfully transformed, and thus these were sent off for sequencing. The sequencing data showed that our construct was correct (see below), so we were able to submit for BioBricking.
Although our sequencing data was good, we realized that we had not induced the lac-repressible promoter, and thus the colonies should not have been fluorescing yet. After doing more research, we realized that the strain of E. coli we were using was not lactose deficient, thus the quorum sensing cascade was being activated by the natural presence of lactose. We ordered a lacI^q strain of E. coli, which are lac deficient, so that we could have complete control of GFP expression. However, when we performed the transformation of the construct into this strain, all of the colonies still fluoresced.
After doing further research, we discovered that the luxPR promoter (BBa_R0062) [2], will induce backward transcription if the luxR protein is present but the AHL molecule is not. In the lac deficient strain of E. coli, AHL is not produced without IPTG induction, but luxR expression is controlled by the constitutive ptet promoter. We hypothesize that since luxR was present in these cells without AHL, the backward transcription from luxPR hindered the function of the terminators that separated the luxR gene from the GFP gene. Thus GFP may have been expressed because the ptet promoter, which is constitutive, was able to affect its expression.
To overcome this issue, in this construct we have added two extra terminators.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1952
Illegal BsaI.rc site found at 2680
Results
Flow Cytometry Results:
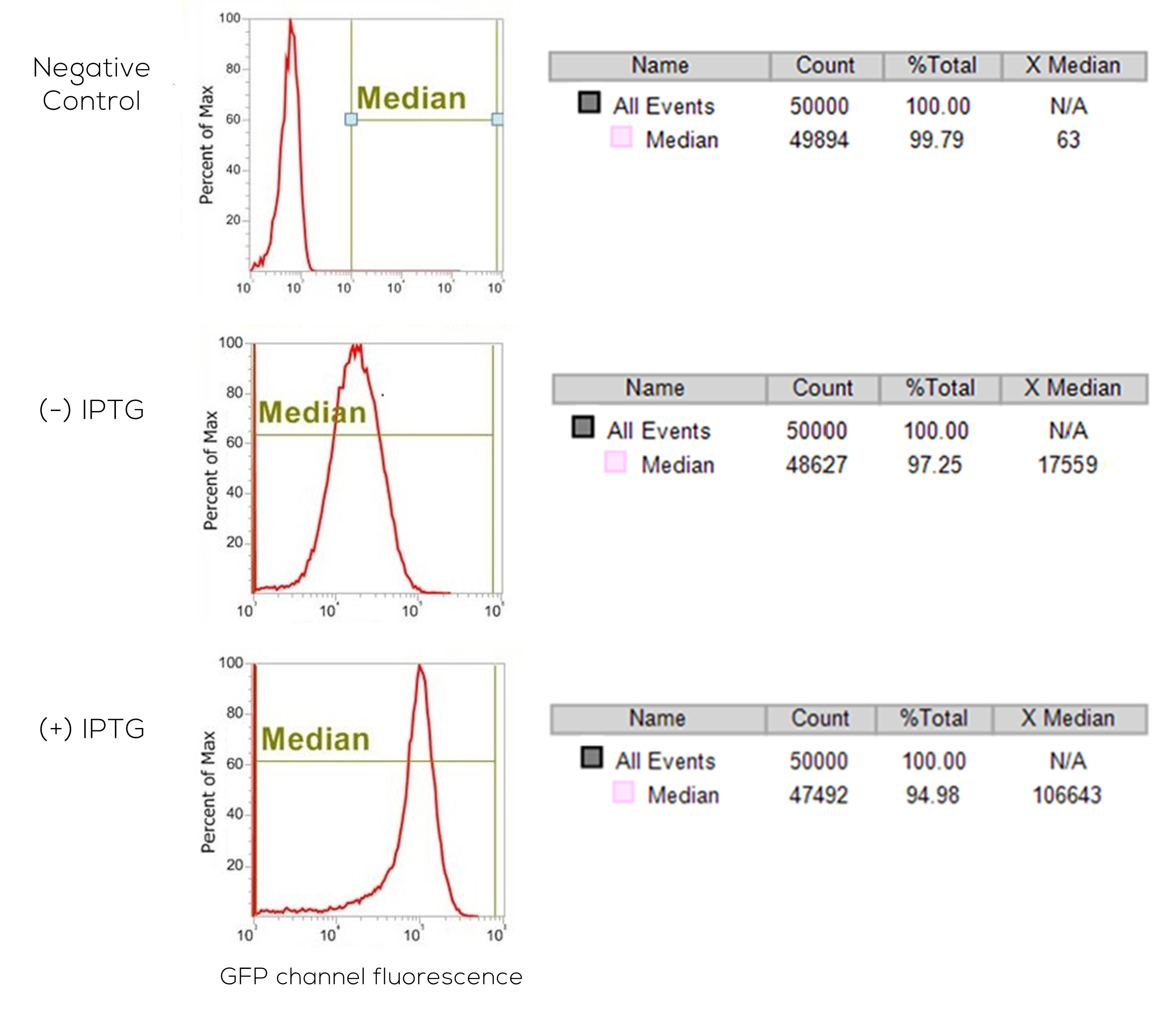
Flow cytometry is a laser-based technology that has a wide-range of uses, including multiparametric cell counting and cell sorting. In this study, we utilized the cell counting ability of the Life Technologies Attune® NxT Acoustic Focusing flow cytometer in order to determine how many of our cells were expressing GFP after being induced with IPTG (a lac analog). The three graphs and tables above represent the flow cytometer data obtained from three samples of lac-deficient E. coli cells. One of the samples was a negative control of LB-cultured E. coli cells (Negative Control), and the other two samples (IPTG-Positive and IPTG-Negative) had the GFP-quorum sensing construct. The IPTG-Positive sample had a ~6X increase in GFP expression over the background IPTG-Negative cells, showing our construct worked correctly.
The “X-Median” seen on the three tables above shows the fluorescence intensity of all of the cells that were counted that were not considered dead (Median). Each sample measured 50,000 cells (Seen under All Events - Count), and then counted all of the cells that were alive (Median - Count), where each sample had between 47,000 and 49,000 live cells. From the data, and as illustrated in the graphs above, we can gather that the fluorescence intensity (X-Median) of the LB only cells was approximately 63 arbitrary fluorescence units (AFU), the IPTG-negative cells was ~17,000 AFU, and the intensity of the IPTG-positive cells was ~107,000 AFU, meaning that the IPTG-positive quorum sensing cells increased fluorescence intensity over 6X the background intensity (IPTG-negative cells). These results indicate that our quorum sensing construct works and greatly increases GFP expression when activated through IPTG-induction. The IPTG-negative cells also expressed some non-negligible degree of GFP, which suggests that the construct could still be improved further. However, the successful proof of concept of our quorum sensing construct is incredibly promising and allows us to proceed forward.
| None |