Part:BBa_K1499253
aeBlue generator with 3 UAG stops + supP tRNA
This part generates aeBlue, a blue chromoprotein. However, it contains 3 amber stop codons that code for leucine through the supP tRNA.
Usage and Biology
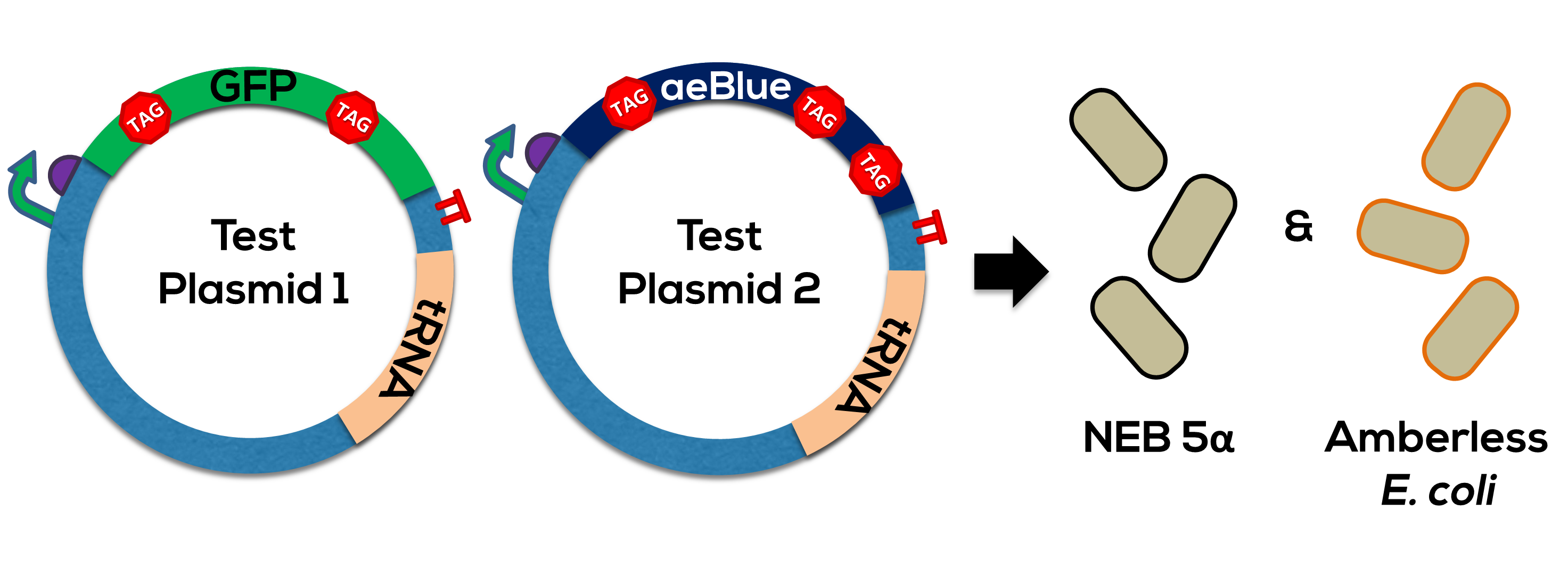
The design of the protein coding region of this part was based off Team Uppsala's 2012 chromoproteins kit, specifically BBa_K864401. Please see this parts page for the biology of this protein. The supP tRNA is from Stanford-Brown-Spelman's 2014 Amberless toolkit, or BBa_K1499251. Together, this generator was used to test the orthogonality of the UAG->Leucine coding (Figure 1).
We have observed that this generator works in amberless E. coli, but not DH5-alpha. Because of the toxicity of the tRNA, the part will be difficult to transform into any strain of bacterium that contains genes with the amber stop codon.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
Verification of Part
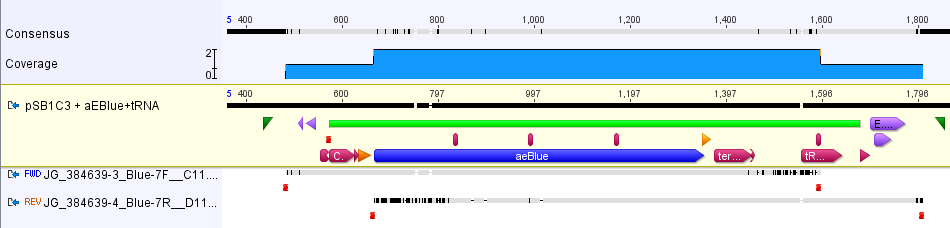
The part was sequence verified in the pSB1C3 backbone before submission to the registry. Two reads, forward and reverse, were obtained using VF2 and VR (Figure 2).
Results
The part has been confirmed to work as expected (Figure 3). The transformation did not work well in DH5-alpha, as seen in the plate on Day 1. The colonies that emerge by day 5 could be due to degradation of the antibiotic and may not represent true transformants. There was no visible expression of the generator in DH5-alpha, but there was expression in amberless cells. This showed that the system could work uniquely in the amberless but indicated it would not work in other cell types.
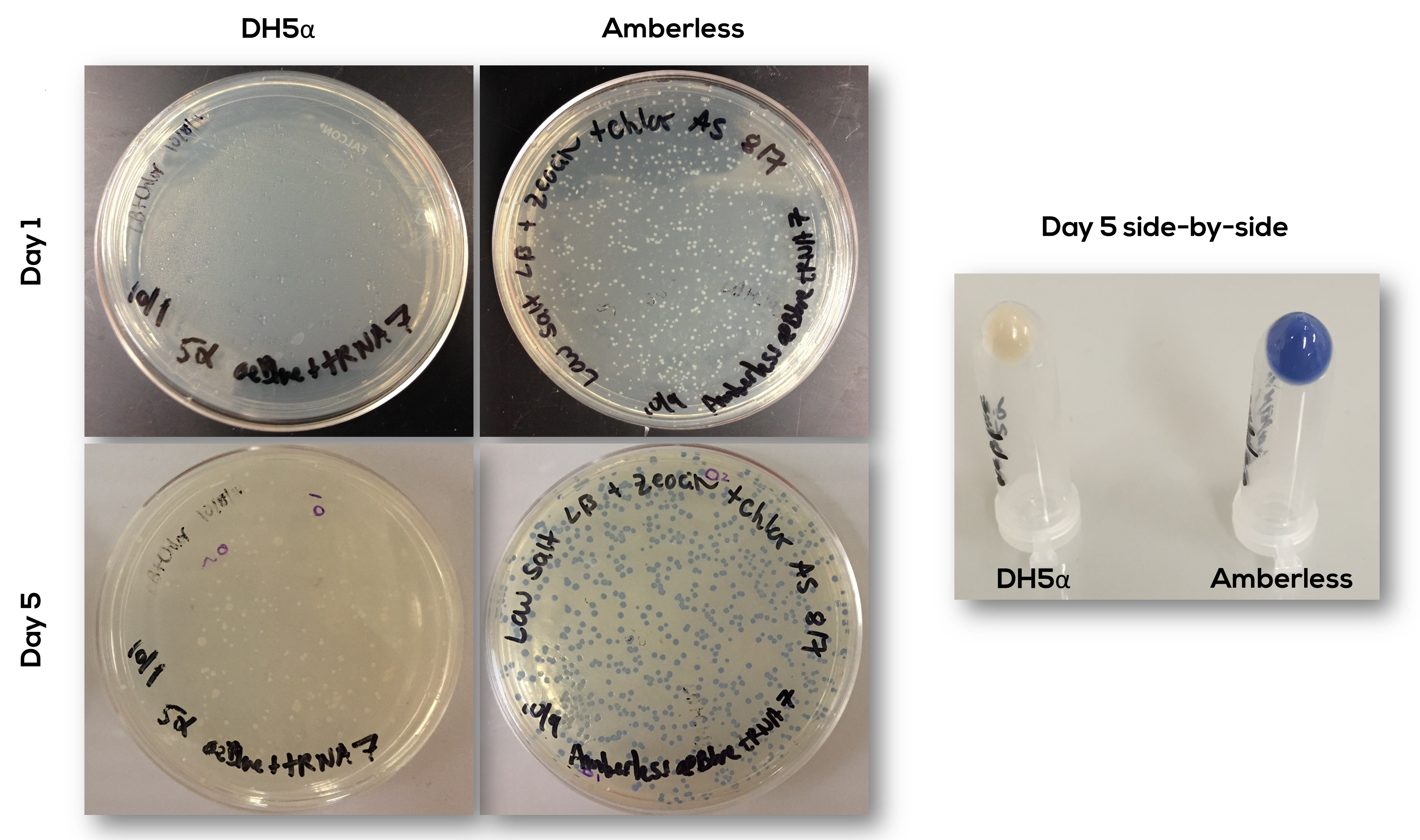
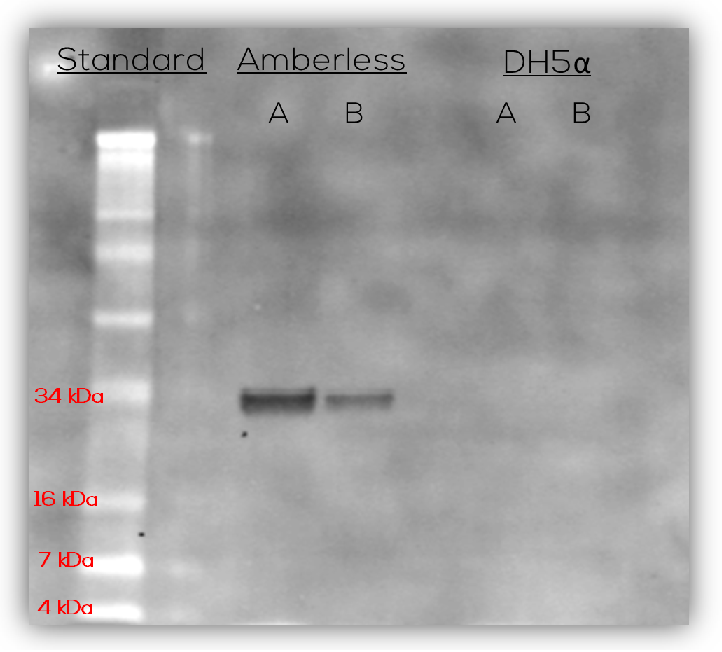
Cells expressed the protein product only with incubation at temperatures of 30C and lower. The first visible blue product appears at Day 2. Finally, a Western blot with an anti-FLAG antibody confirmed that the complete product was produced in amberless but not DH5-alpha cells (Figure 4). There were no incomplete products, or the incomplete products degraded rapidly.
| chassis | Amberless E. coli (C321.ΔA) |