Part:BBa_K1499201:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K1499201
User Reviews
UNIQcf4d63b616cc0b52-partinfo-00000000-QINU UNIQcf4d63b616cc0b52-partinfo-00000001-QINU
In 2019, team Sao_Carlos-Brazil characterized this part expressing it in E. coli as it follows:
Part Characterization
Materials and methods
E.coli TOP 10 strain transformation for part propagation
To transform BBa_K081005, 4 µl of plasmid were incubated on ice for 30 minutes, heat-shocked for 2 minutes at 42°C in water bath. The cell wall was recomposed in test-tube in SOC medium 1ml for 3 hours.
Colonies were identified overnight after incubation at 37°C on solid LB medium with chloramphenicol as a screening method.
In order to transform BBa_K081005, 2 µl of plasmid were incubated on ice for 30 minutes, heat-shocked for 45 seconds at 42°C in a water bath, and later was put back on the ice again. The cell wall was recomposed on LB medium in a microtube for 1 hour. Only a few colonies appeared after incubation for 36 hours at 37°C in solid LB medium with chloramphenicol as a screening method.
Miniprep
To extract both part's plasmidial DNA, a Cellco kit was used. Isolated TOP10 E. coli colonies were chosen from the previously obtained plates that were cultivated overnight in 5ml of LB medium with chloramphenicol. DNA was obtained from the selected colonies.
Digestion
BBa_K1499201 part was digested using 1.8 µl of DNA (554 ng/µl). Digestion was carried out with 1µl XbaI (NEB) and 1µl PstI (NEB), µl of NEBuffer 2.1, 1µl of BSA and 41. µl of MilliQ water. Incubation was carried out at 37°C.
BBa_K081005 was digested using 3µl of DNA (331 ng/µl), 1 µL of SpeI (NEB), 1µL of PstI (NEB), 5µl of NEBuffer 2.1, 1µL of BSA and 39 µl of MilliQ water. Incubation was carried out for 1h at 37°C.
After an electrophoresis, the digested DNA fragmenst were purified from the agarose gel.
BioBricks ligation
The ligation step was carried out with 0.7 µL of digested BBa_K081005 as the backbone, 3.9 µL of digested BBa_K1499201 as insert, 2 µL buffer solution, 1 µL T4 Ligase (Thermo Fisher Scientific) and 12.4 µL MilliQ water. The reaction was incubated for 5 days at 18°C. The control consisted of a reaction without the insert and the same reagent volumes.
Transformation
Competent BL21 E. coli cells were transformed utilizing 20µL from the resulting solution from the ligation step. The transformation was incubated 30 minutes on ice, and the cells were heat-shocked for 2 minutes at 42°C. Cell wall recomposition was carried out in a microtube tube for 1 hour in 1ml of SOC medium. Colonies were confirmed after overnight incubation in solid LB medium with chloramphenicol, at 37°C. Control reaction followed the same parameters, but without the plasmid.
Another transformation reaction was carried out, with the same parameters but with only BBa_K01005 as insert. This reaction resulted in E. coli that was later used as a control for the phenotypic test.
LB medium
The E. coli was cultivated in Luria-Bertani medium (LB), composed of tryptone1% (p/v), yeast extract 0,5% (p/v) and NaCl 0,5% (p/v). The pH was adjusted to 7,5. In order to make solid medium, agar 2% was added.
Saline solution
For cell dilutions, 0.9% (w/v) NaCl solution was used.
Expression test
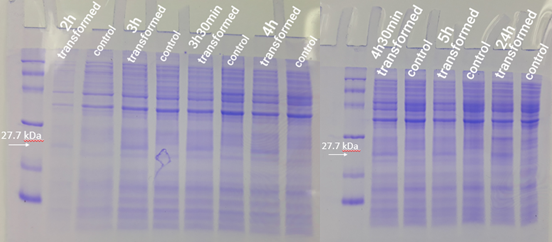
2.5 mL of transformed bacteria were inoculated into 250 mL of LB medium and incubated at 37°C at 120 rpm. Bacteria without the plasmid were also inoculated and used as a control group. Then, 500 mL aliquots were taken from each of the strains (transformed and not transformed) at 1h, 2h, 3h, 3h30min, 4h, 4h30min, 5h, and 24h, counted from the incubation. To follow the bacterial growth rate, the optical density was also inferred at each period. Aliquots removed were centrifuged for 1 minute at 16000g. Then, the formed pellet was resuspended in freshly prepared running buffer (800 µl Loading Buffer 4x, 200 µl DTT). For the first point (1h period), 30 µl of buffer was used for resuspension. For the other points, 60 µl of buffer was used. Then the mixture was boiled for 5 minutes. Test analysis was performed using 15% SDS PAGE gel, applying equalized sample volumes.
Preparation of phenotype test plates
Transformed and control BL21 E. coli were inoculated into a test tube containing 2 mL of liquid LB medium. Incubation was carried out at 37°C, 150 rpm, for 24h. The extended incubation period was intended to guarantee that radiation tests would be performed with organisms in the stationary phase.
The plates were homogenized by shaking for 3 minutes. Then, the first aliquots were taken from each plate to make up the control point (without irradiation). Each aliquot corresponds to a volume of 200 µl transferred to a sterile microtube. The Petri dishes containing the samples were uncapped and subsequently exposed to the first dose of UVC radiation (254 nm) in USP São Paulo Astrolab’s irradiation chamber of approximately 50 J/cm², as shown in Figure 2. After this exposure, the second aliquot was collected. (200 µL) of each plate. Thus the following doses of 100 J/cm², 200 J/cm², 400 J/cm², and 800 J/cm² were continued. At each exposure point, an aliquot was taken from each of the plates (control and transformed strain). Samples from each point were exposed to the corresponding radiation doses for a period equivalent to that required for the irradiation equipment to reach the corresponding energy level. This period is equivalent to a few seconds. For example, the equipment took about 5 seconds to reach 50 J/cm². To reach 100 J/cm², the equipment took about 8 seconds.

Plating
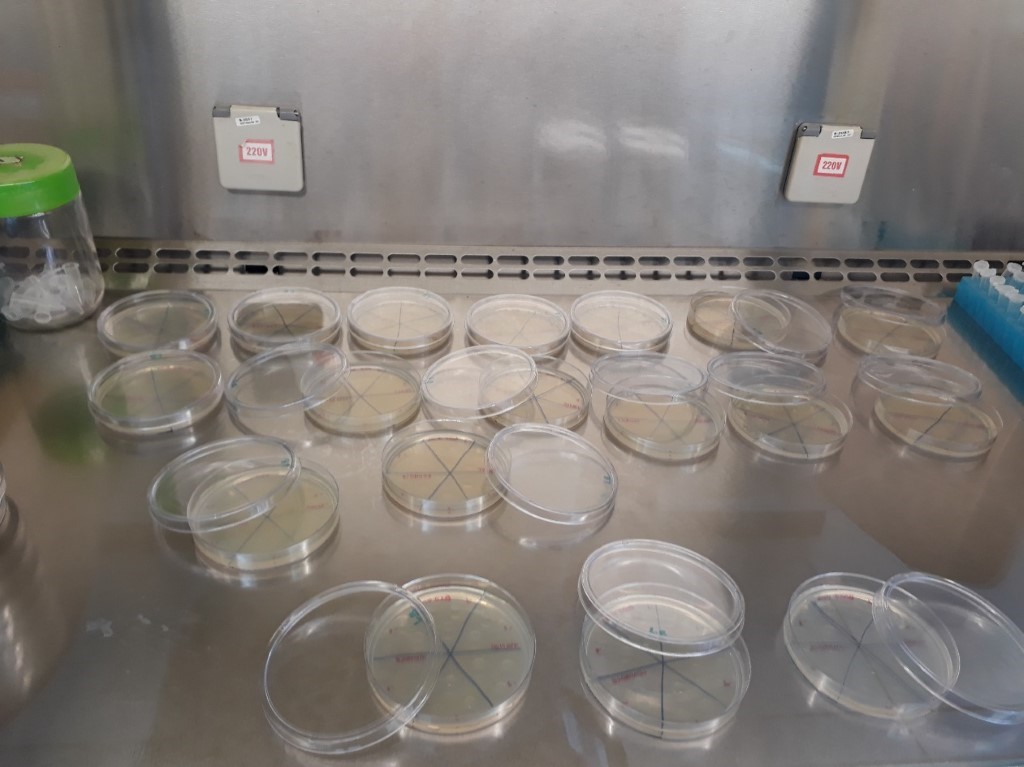
After irradiation of the samples, each irradiation point aliquot was inoculated in Petri dishes containing 20 mL of solid LB medium by a drop plating. Plating was performed in triplicate and samples were diluted in 3 distinct concentrations. The plates were divided into 6 parts to hold the 3 distinct dilutions of each of the strains, as can be seen in figure 3.

Samples from the control point (without irradiation) were diluted to concentrations of 10-3, 10-4 and 10-5. Serial dilution was done by adding 50 µl of sample into a sterile microtube containing 450 µl of saline solution. From each dilution, 3 drops of 10µl each were transferred to the plates containing solid LB medium (Figure 4).

For plating the irradiated samples (50 J/cm², 100 J/cm², 200 J/cm², 400 J/cm², and 800 J/cm²) dilutions of 10-2, 10-3 and 10-4 were used. As the irradiated sample was already in the 10-2 dilution, the aliquot itself was taken directly from the irradiated Petri dish to performing the drop. The 10-3 dilution was performed by adding 50 μl of the original sample (concentration 10-2) in sterile microtube containing 450 μl of 0.9% saline solution and the 10-4 dilution was performed by adding 50 μl. µL of 10-3 dilution in sterile microtube containing 450 µL of 0.9% saline solution. After preparing the dilutions, 3 drops of 10 µL from each dilution were added to Petri dishes containing solid LB medium.
The plates were left open in the Laminar flow cabinet until the drops dried completely (figure 5). The plates were then incubated at room temperature for a total of 40 hours.
Results
The expression test presented positive results for the expression of the desired protein. In figure 6, it is possible to visualize bands of expected size (27.7 kDa) in the transformed strain samples, while in the control group there is no band of the mentioned size. This demonstrates that D. radiodurans Uracil DNA glycosylase protein was successfully expressed by the strong constitutive promoter associated with a strong RBS. This provided support for phenotypic testing, with irradiation of samples at progressive doses of ultraviolet C radiation.
Phenotype testing was inconclusive up to 100 J/cm², which means that it was not possible to ascertain whether the foreign protein provided some protection against UV radiation. However, it is likely that no resistance was induced between 50 and 100 J/cm². It was conclusive though that beyond this irradiance, very few CFUs were observed, meaning that D. radiodurans’ uracil DNA Glycosylase does not provide protection against UVC when cells are irradiated with such a large dose of radiation. The following images and graphic illustrate our results, with no statistical significance encountered for control and transformed groups after a Two-Way ANOVA test.

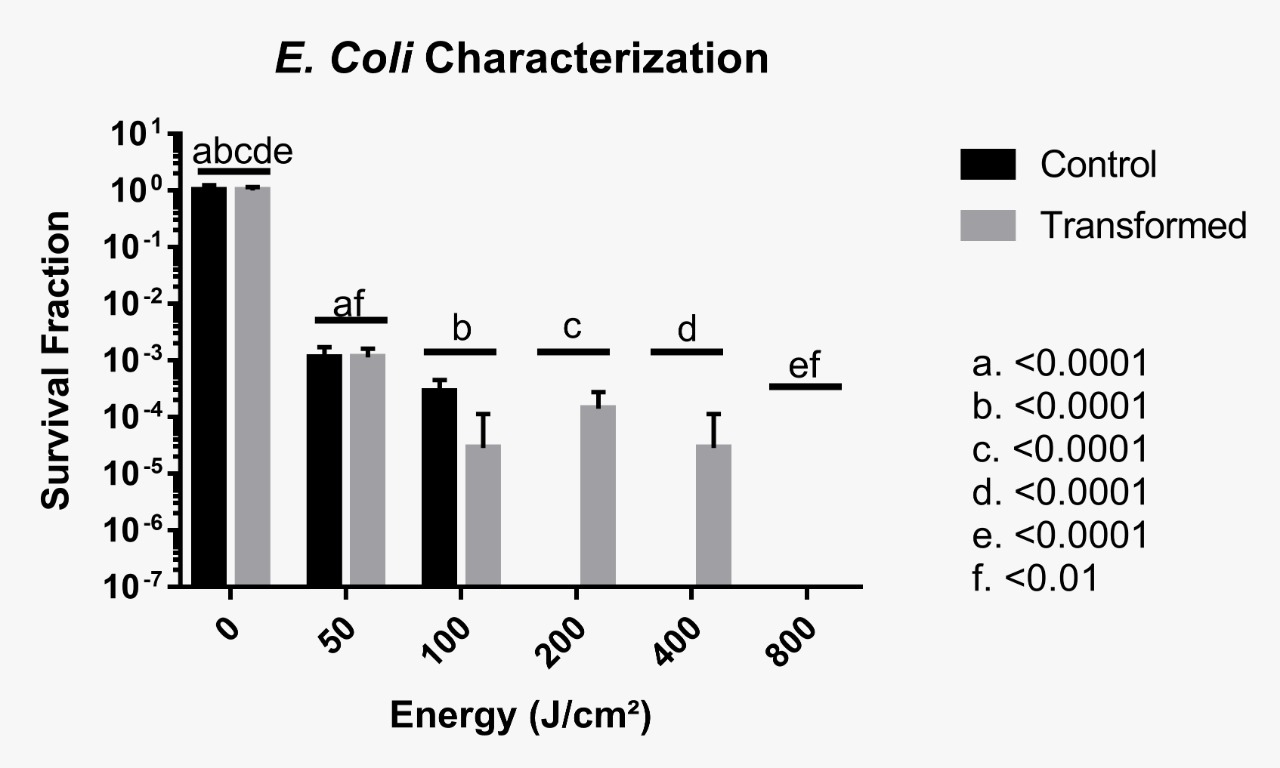
It is important to emphasize that the results presented by cells irradiated at 100 J/cm² conflict with the others, with a higher survival rate in the control group when compared to the transformed group. A hypothesis that may explain this result is an experimental error, as there may have been an unreasonable exchange of strains during plating. The following image depicts the triplicate plates of cells irradiated at 100 J/cm². Another possible explanation is that the protein is somewhat toxic to E. coli cells, or maybe that this irradiance level cannot be sensitive enough to discriminate differences between control and transformed groups.

The team conducted three additional tests in order to confirm these results, but they were all unsuccessful. For the first attempt, we irradiated the cells using a crosslinker (Stratagene’s UV Stratalinker 1800), but there were no survivors from the first dose onwards. We believe the irradiance became chronic and not acute because despite irradiance levels are significantly lower than Astrolab’s irradiation chamber, the exposure time was considerably larger. Furthermore, the crosslinker is a smaller sealed environment, which could have promoted a larger ozone concentration in the interior of the machine. This may have resulted in accelerated death due to ozone toxicity to cells. The second attempt was conducted in Astrolab’s irradiation chamber and was identical to the first experiment but with extra aliquots for 25 and 75 J/cm². However, for unknown reasons, not radiated transformed cells did not even grow on solid LB medium, therefore rendering this experiment lost. A third and final attempt was made using the UVC lamp of our São Carlos lab LBEst, but the results were too inconclusive due to problems in quantifying the irradiance doses and CFU counting.
For future experimentation, we recommend using lower irradiation doses given that beyond 100 J/cm², it was clear that the protein did not confer resistance to bacterial cells. This might be the case for lower doses but this is yet to be verified.
