Part:BBa_K1499201
Deinococcus radiodurans uracil-DNA glycosylase 1
This part encodes a uracil-DNA glycosylase found in D. radiodurans, which protects cells from radiation-induced DNA damage. In the literature, it is referred to as DR0689 and detailed protein features can be found on its [http://www.ncbi.nlm.nih.gov/protein/NP_294412.1 NCBI Protein] page.
Usage and Biology
Deinococcus radiodurans is a highly radiation-resistant bacterium that can survive after up to 12,000 Gy (absorbed radiation dose, Gray) of radiation. The unique genes that allow it to better handle radiation exposure may provide ways to make E. coli and other organisms more resistant for applications of synthetic biology in tough environmental conditions, like space. D. radiodurans is known to survive both ionizing and UV radiation.
One gene that contributes to this phenotype is a family 1 uracil-DNA glycosylase (UDG), hereafter called uracil-DNA glycosylase 1 (UDG1), which catalyzes the excision and repair of cytosine->uracil mutations that occur following UV exposure [http://www.ncbi.nlm.nih.gov/pubmed/14706350 (1)][http://www.ncbi.nlm.nih.gov/pubmed/10946227 (2)]. The enzyme can remove uracil from U:G and U:A pairs in both double-stranded and single-stranded DNA. Out of the four families of UDGs, family 1 UDGs are found in E. coli, humans, and DNA viruses [http://www.ncbi.nlm.nih.gov/pubmed/11807060 (1)].
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 720
- 1000COMPATIBLE WITH RFC[1000]
Characterization
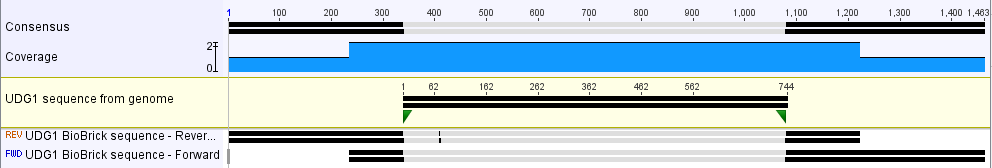
Verification of Part
The part was sequence verified in the pSB1C3 backbone before submission to the registry. Two reads, forward and reverse, were obtained using VF2 and VR (Figure 1).
Results
Functional data are in the process of being obtained. Transformation of the part ligated to a constitutive promoter and medium RBS failed twice, suggesting that it may be toxic if it is highly expressed.
References
1. Sandigursky, M et al. (2004) Multiple uracil-DNA glycosylase activities in Deinococcus radiodurans. DNA Repair (Amst) 3(2):163-9. PMID: [http://www.ncbi.nlm.nih.gov/pubmed/14706350 14706350].
2. Pearl, LH (2000) Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res. 460(3-4):165-81. PMID: [http://www.ncbi.nlm.nih.gov/pubmed/10946227 10946227].
//chassis/prokaryote/ecoli
| biology | Deinococcus radiodurans |
| protein | uracil-DNA glycosylase |