Part:BBa_K1390019
Complete sMMO of Methylococcus capsulatus
The part consists of the six subunits of methane monooxygenase (MMO). The expression is under control of lac+pl promoter. MMO converts methane to methanol.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NotI site found at 1926
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1236
Illegal BamHI site found at 1270 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1883
Illegal NgoMIV site found at 2719
Illegal NgoMIV site found at 2918
Illegal NgoMIV site found at 4705
Illegal NgoMIV site found at 4883
Illegal AgeI site found at 2130 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 4844
Illegal BsaI site found at 4909
Illegal BsaI.rc site found at 245
Illegal BsaI.rc site found at 383
Illegal BsaI.rc site found at 800
Illegal BsaI.rc site found at 1949
Illegal BsaI.rc site found at 3995
Illegal BsaI.rc site found at 4952
Contents
Biology and Usage
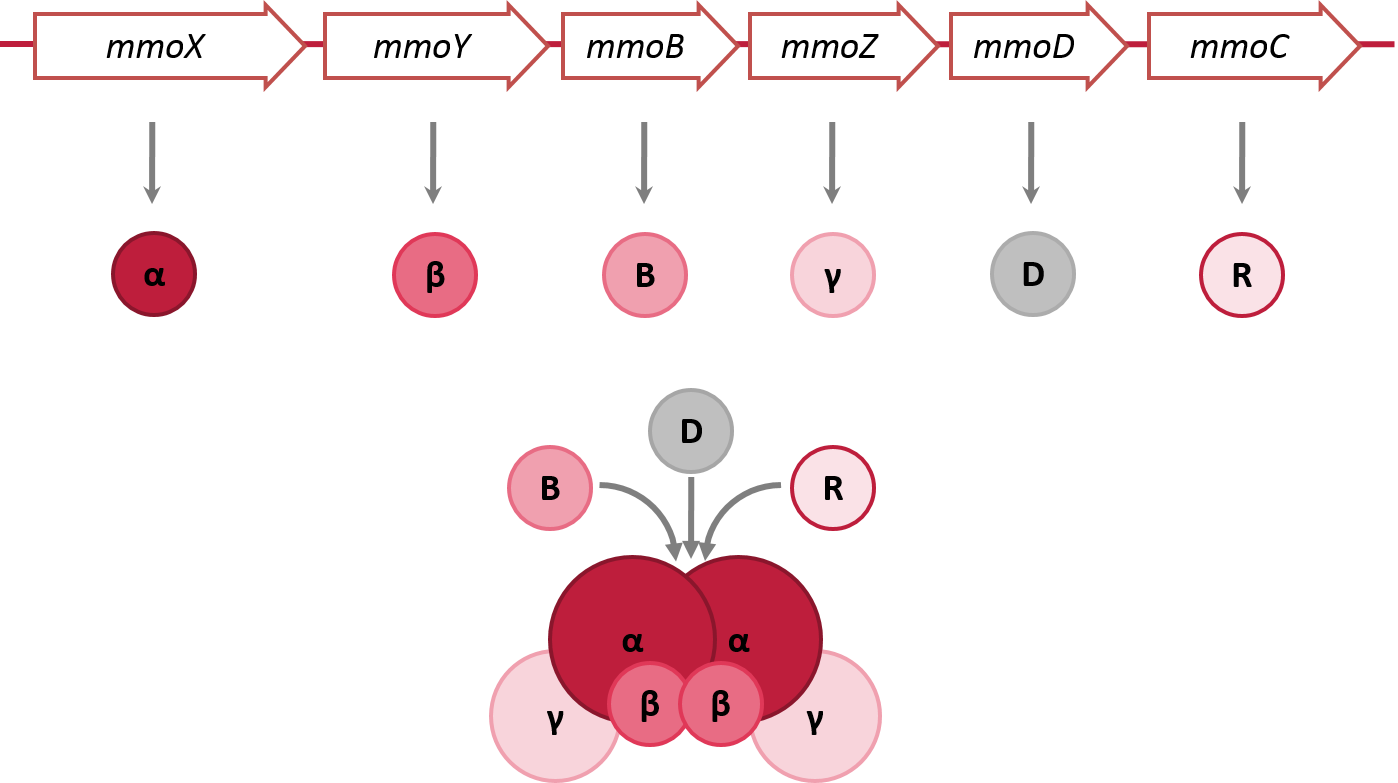
The soluble methane monooxygenase (sMMO) from Methylococcus capsulatus is an enzyme complex capable of oxidizing the greenhouse gas methane to methanol. The complex consists of three main components all of which are required for proper enzyme function. The hydroxylase component itself is made up of the three subunits α, β and γ which are arranged as a heterodimer (α2β2γ2) (Figure 1). The α subunit houses the active site of the enzyme where the hydroxylation of methane takes place. The reductase component MMOR oxidizes NAD(P)H and shuttles the electrons to the hydroxylase. The electron transfer is facilitated by a [2Fe-2S] cluster and an FAD cofactor bound to the reductase. The third component, the regulatory protein MMOB, seems to have a function in modulating electron transfer because it binds to the same site of MMOH as MMOR [1]. Another open reading frame within the mmo operon encodes MMOD whose function has not yet been clearly determined [2,3]. Despite this uncertainty we included all components of the mmo operon in our strategy.
The sMMO has the potential to conquer three major concerns of the industrialized economy: pollution, shortage of energy and, of course, global warming.
As the sMMO is able to degrade a broad variety of substrates such as one of the major water pollutants, the industrial solvent trichlorethylene (TCE) [4], our project can serve as a foundation for a new approach of biological water treatment or environmental care in general.
One could also exploit the ability of the sMMO to convert methane to methanol: Methane can serve as an energy source, but due to its gaseous state it is very difficult to transport compared to liquid methanol. So far methane is chemically converted to methanol for transportation and later on further converted to components of diesel fuel and gasoline or to propylene and ethylene, precursors of important chemical substances. Using the sMMO to oxidize methane is a much more economical and eco-friendly approach to utilize methane as a resource for energy production [5]. This wide range of usability makes the sMMO so interesting for synthetic biology.
The major problem of the use of methanotrophs as MMO suppliers is that they are very difficult to cultivate. They cannot live without methane and grow very slowly. Because of the slow growth it is not possible to isolate sufficient amounts of the enzyme. The scale-up process is not economical enough for industrial application.
Design and Coexpression
BBa_K1390019 consists of six different subparts corresponding to the subunits of the original sMMO. We put each of the genes under the control of the inducible lacl promoter (R0011) which has a low leakiness and is easily inducible with IPTG. This strong, frequently used promotor is well characterised in the iGEM registry and is reported to be well functioning. We also added the extensively documented weak ribosome binding site (B0032) with a strength of 33.96% compared to B0034 to diminish the formation of inclusion bodies and a double terminator (B0015). Compared to the natural cluster of M. capsulatus BBa_K1390019 does not include the naturally occuring transposases and hypothetical proteins. However, the arrangement order of the subunits is the same as in the host organism. For expression we decided to use pSB1A3 because of its ampicillin resistance (in contrast to the chaperone plasmid which carries a chloramphenicol resistance cassette). To ensure proper folding of the BBa_K1390019 components in E. coli JM109 we coexpressed it with chaperones (GroES, GroEL). The TaKaRa Clontech Chaperone Plasmid Set proved to be appropriate for this as it contains a chaperone plasmid encoding GroES and GroEL ([http://2014.igem.org/Team:Braunschweig/Results-content#chaperone_table chaperone plasmid C2]) which can be coexpressed with BBa_K1390019 [6]. Synthesis of the part-encoded proteins can be induced as soon as the chaperones are operational.
Part Analysis
Growth statistic
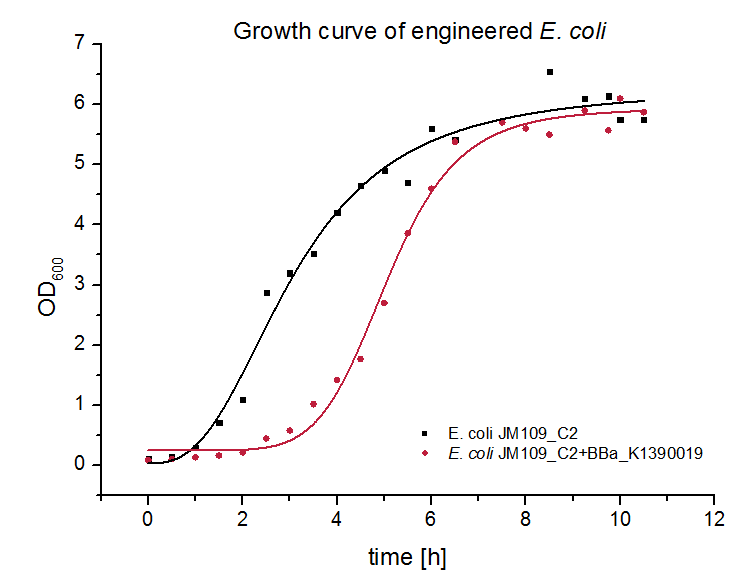
To investigate whether BBa_K1390019 or the coexpressed chaperones inhibit the growth of E. coli we performed growth analysis (Figure 2).
When BBa_K1390019 is coexpressed with the chaperone plasmid the growth is limited for a short period of time in comparison with E. coli JM109 transformed solely with the chaperone plasmid. This limited growth could be caused by a greater replication stress. However, when approaching the stationary phase both systems grow to the same OD. Obviously BBa_K1390019 does not cause any toxic effects for E. coli.
Expression Analysis
To detect whether the subunits are synthesized in soluble form or in inclusion bodies they were provided with a 6xHis-tag, as shown for BBa_K1390007 - BBa_K13900012. Thus it is possible to check the expression of each subpart individually. We analyzed the expression by Western Blot. We separated the soluble fraction and the inclusion body fraction of each subpart after expression of BBa_K139007 - BBa_K1390012 with chaperone plasmid 2 . With the exception of BBa_K1390008 (Figure 3a) all subparts were expressed with the aid of GroES and GroEL encoded on the chaperone plasmid (Figure 3b).
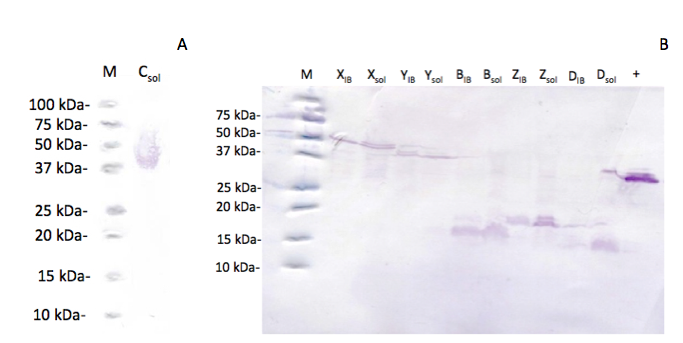
Using the chaperone plasmid resulted in expression of all subparts. Although a substantial amount was still detected in inclusion bodies these results indicate soluble expression of all His-tagged components of the enzyme complex (FIGURE 3b). In conclusion, all subunits of sMMO can be synthesized in E. coli JM109 in the soluble fraction. BBa_K139014 does not need any folding proteins, whereas all other subparts need to be coexpressed with the chaperones GroES, GroEL [6]. Since all subparts can be expressed in the soluble fraction it can be assumed that the composite part consisting of all these subparts can also be expressed correctly. Furthermore, the activity assay suggests correct assembly of the subunits.
Activity Assay
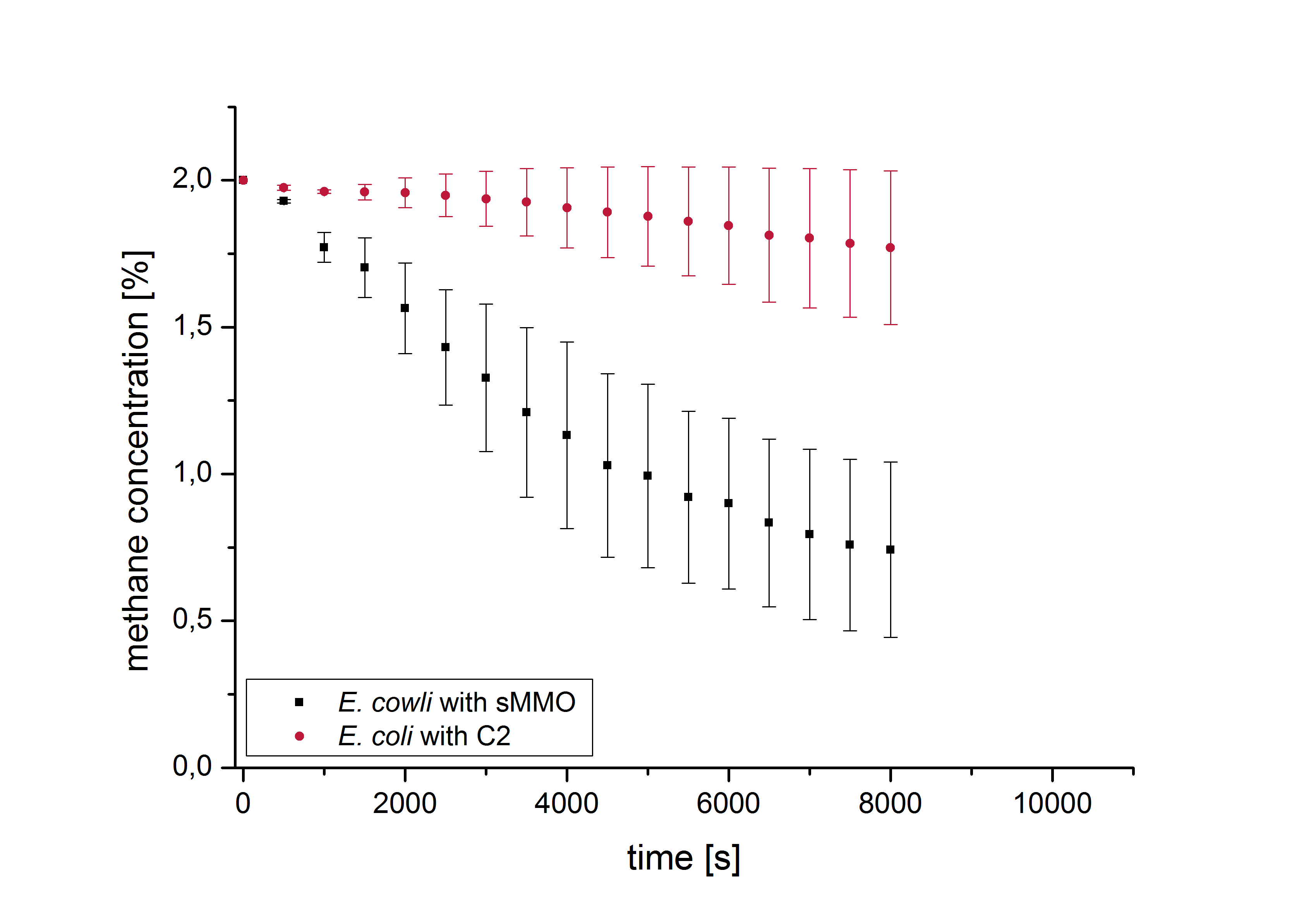
To determine the functional expression of sMMO in our modified bacterium, activity tests were performed by measurement of the methane concentration. These experiments were carried out in a converted anaerobic jar using an MQ-4 Semiconductor Sensor for Natural Gas (Henan Hanwei Electronics Co., Ltd.). The jar was filled with a defined atmosphere of 2% methane and 98% air. The experiments were performed with M. capsulatus and E. coli transformed with BBa_K1390019 and chaperone plasmid 2 to compare the degradation rate of our part with that of the natural enzyme complex. The measurements were performed three times with bacteria cultures of an OD600 value of 4 in NMS medium [7,8] and a wet biomass of 0.5 g and a mean value was calculated. E. coli JM109 transformed with chaperone plasmid 2 served as a negative control which ought not be able to degrade methane. The methane degradation rate was measured three times for this strain as well as for E. coli transformed with the plasmid carrying BBa_K1390019. A mean value was calculated from these three measurements. As expected, hardly any methane was degraded in the negative control. In contrast, the methane concentration in the jar decreased from 2% to about 0,75% within 8,000 seconds (≈133 min) indicating that E. coli transformed with BBa_K1390019 and chaperone plasmid 2 is indeed able to degrade methane (Figure 4). This demonstrates that it is possible to produce all subunits in the soluble fraction and that they can assemble to form a functional complex. The resulting recombinant sMMO/BBa_K1390019 shows a substantial enzyme activity.
Summary
- BBa_K1390019 does not inhibit the growth of E. coli
- metabolism is not influenced
- BBa_K1390019 is synthesizable in the soluble fraction by use of several chaperones.
- we recommend the use of GroEL/GroES provided by Takara (Takara Bio Inc., Japan)
- BBa_K1390019 is able to degrade methane
For more detailed information on this part check the [http://2014.igem.org/Team:Braunschweig/Results results page] of iGEM Team Braunschweig 2014..
- 2021 OkState update:
- We created two new composite parts BBa_K4884006 and BBa_K4884007 based on this composite.
- We codon optimized all mmo genes as the original sequences were from M. capsulatus.
- We have also replaced the promoter with an Arabinose inducible promoter with the the reasoning that since the Takara Biotech Chaperone plasmid needed to be co-transformed, and since the chaperones are inducibled by arabinose, it might be easier to have one inducible system to turn on the expression of plasmid in E. coli.
- In the future, we believe another improvement would be to mutate specific residues of the mmo genes to improve activity. According to this publication https://www.biorxiv.org/content/10.1101/2021.08.05.455234v1 by Bennett et al, the authors was not only successful in expressing a functional sMMO from M. capsulatus in E. coli, they went another step further and identify mutations that increased the enzyme activity. Specifically they found that mutations mmoB (Y139S), mmoX (V23G), and mmoZ (R70E) along with mutation in GroEL2 of M. capsulatus (N409G) increased the methane oxidate rate significantly. Another interesting finding of the paper is that the group demonstrated that two chaperones are still required for proper folding, but they were able to use the GroES EL2 of M. capsulatus as one of the chaperone rather than using two E. coli chaperone proteins.
- Please visit the 2023 iGEM Team OKstate page for more information about our updated Parts page.
Functional Parameters
| biology | rrnBT1-T7TE |
| chassis | E.coli |
| direction | Forward |
| efficiency | 0.3 |
| forward_efficiency | 0.984[CC] 0.97[JK] |
| negative_regulators | 1 |
| positive_regulators | -NA- |
| reversed_version | 2146 |
| reverse_efficiency | 0.295[CC] 0.62[JK] |
References
[1] Wang W, Iacob RE, Luoh RP, Engen JR, Lippard SJ (2014) Electron Transfer Control in Soluble Methane Monooxygenase. J Am Chem Soc 136:9754-62
[2] Merkx M, Lippard SJ (2001) Why OrfY? Characterization of MMOD, a long overlooked component of the soluble methane monooxygenase from Methylococcus capsulatus (Bath). J Biol Chem 277:5858-65
[3] Semran JD, Jagadevan S, DiSpirito AA, Khalifa A, Scanlan J, Bergman BH, Freemeier BC, Baral BS, Bandow NL, Vorobev A, Haft DH, Vuilleumier S, Murrel JC (2013) Methanobactin and MmoD work in concert to act as the “copper-switch” in methanotrophs. Environ Microbiol 15:3077-86
[4] Brussea GA, Tsien HC, Hanson RS, Wackett LP (1990) Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation 1:19-29
[5] Tinberg CE, Lippard SJ (2011) Dioxygen Activation in Soluble Methane Monooxygenase. Acc Chem Res 44:280-8
[6] Chaperone Plasmid Set Product Manual, TaKaRa Bio Inc. 2014
[7] [http://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium1179.pdf http://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium1179.pdf] Date: 17.10.2014
[8] [http://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium632.pdf http://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium632.pdf] Date: 17.10.2014
//ribosome/prokaryote/ecoli
//chassis/prokaryote/ecoli
//direction/forward
//regulation/constitutive
| biology | rrnBT1-T7TE |
| chassis | E.coli |
| direction | Forward |
| efficiency | 0.3 |
| forward_efficiency | 0.984[CC] 0.97[JK] |
| negative_regulators | 1 |
| positive_regulators | |
| reversed_version | 2146 |
| reverse_efficiency | 0.295[CC] 0.62[JK] |