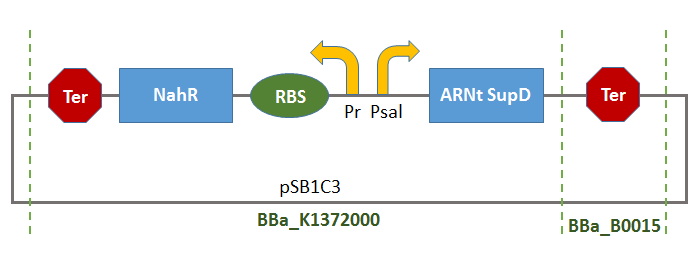
Part:BBa_K1372001
Salicylate Inducible Suppressing System
This Biobrick is the result of the assembly of two Biobrick:
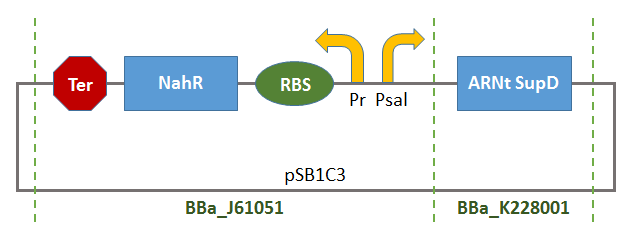
- BBa_K1372000:
- BBa_B0015: Double terminator consisting of BBa_B0010 and BBa_B0012.
Thus, there will be a terminator of the transcription of tRNA-SUPD gene (missing in BBa_K1372000).
This Biobrick could thus be used as control system of the production of tRNA-SUPD depending on the concentration of a substrate (salicylate) gradually consumed by the organisms in the growth-medium. Consequentially, the production of tRNA-SupD will gradually peter out.
An example of use of this control system is described there: [http://2014.igem.org/Team:Paris_Saclay/Project/Salicylate_Inducible_System Salicylate Inducible System]
Characterization of BBa_K1372001
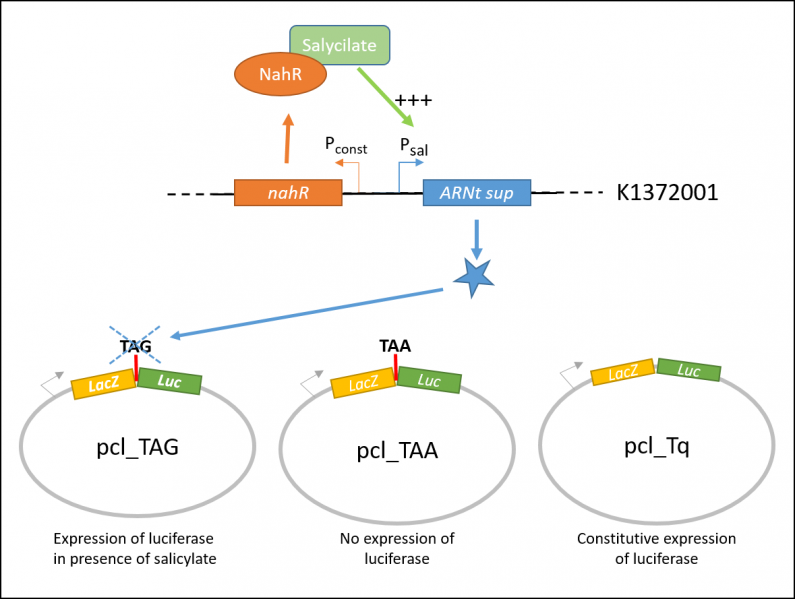
In order to characterize the biobrick, the team iGEM Paris Saclay 2016 tested the salicylate inducible suppressing system (BBa_K13372001) on three different constructions, contained in the plasmid pcl99:
- TAA: LacZ and Luc coding sequences in the same open reading frame, separated with an ochre stop codon.
- TQ: LacZ and Luc coding sequences in the same open reading frame.
- TAG: LacZ and Luc coding sequences in the same open reading frame, separated with an amber stop codon.
Each condition was tested under three different salicylate concentrations. In order to achieve that, both measurements of Beta-Galactosidase and Luciferase activities were performed on bacteria cultures [Fig. 1].
The experiment was conducted on three sets of cultures of bacteria:
- TAA: BL21|pSB1C3_BBa_K1372001 and pcl_TAA
- TQ: BL21|pSB1C3_BBa_K1372001 and pcl_Tq
- TAG: BL21|pSB1C3_BBa_K1372001 and pcl_TAG
Each of those sets of culture were incubated with three different salicylate concentrations: 0, 30µM and 1mM.
Three clones (clones 1, 2 or 3) were tested for each condition, with three different salicylate concentrations (0, 30µM or 1mM), with in addition a negative control sample.
pcl_TAA construction contains a TAA stop codon between LacZ and Luc. This codon is not recognized by the supD suppressor t-RNA : no luciferase activity is expected. pcl_Tq construction does not contain any stop codon : the luciferase activity is expected to be at the maximal. pcl_TAG contains the TAG codon recognized by supD suppressor t-RNA : the expression of luciferase should be inducible by salicylate.
The luciferase luminescence is expected to vary depending to the different constructions conditions and to salicylate concentrations, instead of the Beta Galactosidase activity, which will remain constant. Thus luciferase data were normalized with those from Beta Galactosidase and our results are expressed as the Luciferase/Beta-Galactosidase activity. This ratio is independent of the level of transcription, initiation or mRNA stability.
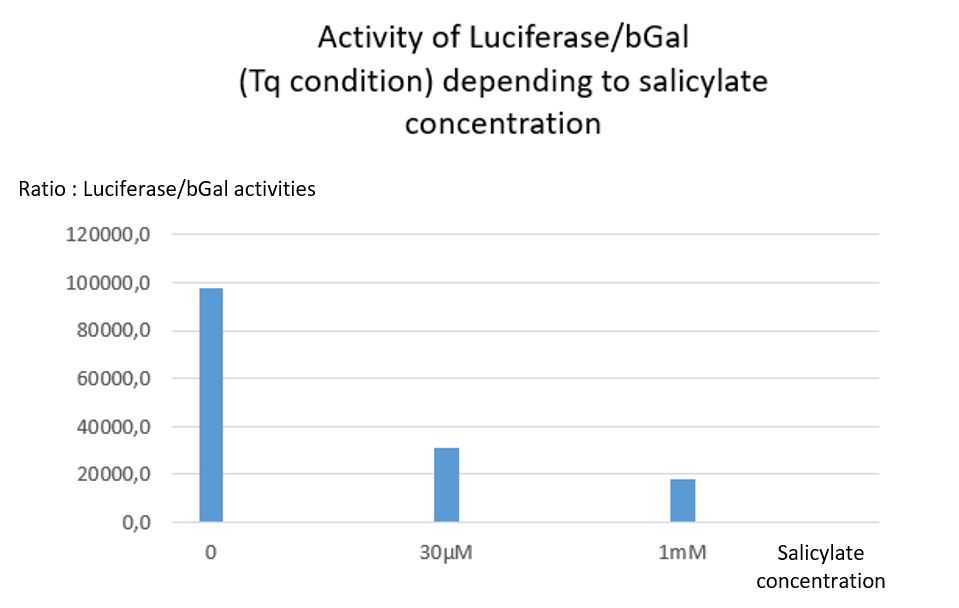
The Tq plasmid does not contain any stop codon between LacZ and Luc. Thus, no matter the salicylate concentration, both Luciferase and Beta Galactosidase activities are supposed to be detected. As expected a high level of Luciferase/bGal activity is observed, but the ratio decreases when salicylate concentration increases [Fig. 2]. Indeed, both activities of Luciferase and bGal drop from 30µM of salicylate, but luciferase activity was more affected by the salicylate than bGal one. One may hypothesize that salicylate inhibits differently both reporter proteins activities, with a stronger inhibition of luciferase activity. We cannot determine whether this inhibition is due to a physiological consequence onto bacteria metabolism or occures after protein extraction.
In order to read the next results, we calculated a readthrough percentage, by doing a ratio between luciferase/bGal from TAA or TAG constructs and luciferase/bGal from TQ. Thus, we obtain a readthrough percentage.
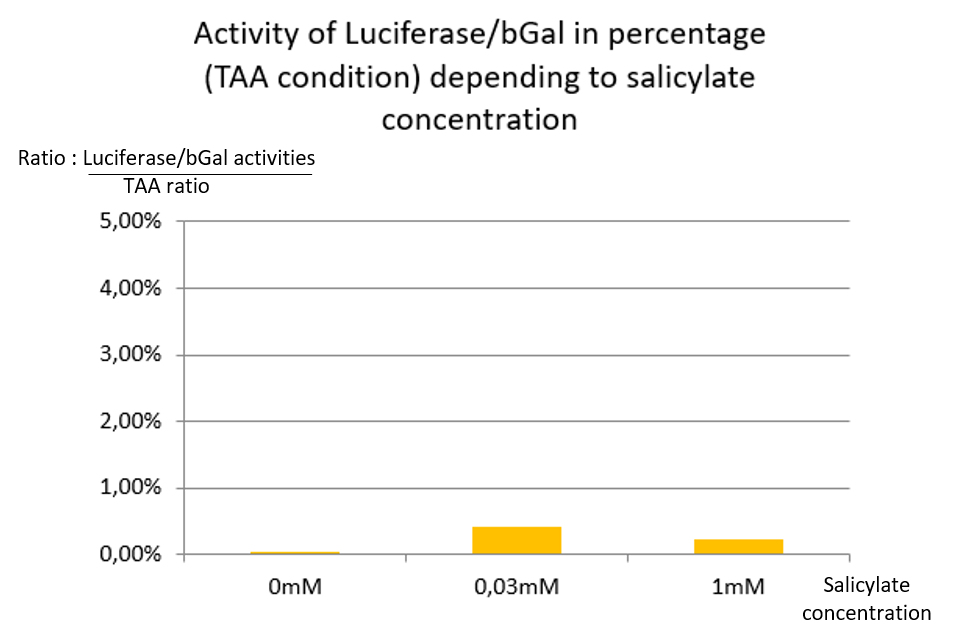
In TAA condition, regardless of the salicylate concentration, there is no significant Luciferase activity, so the ratio remains very low at any concentrations [Fig. 3].
We conclude that supD suppressor tRNA is very specific of the TAG codon and has no impact on the TAA stop codon.
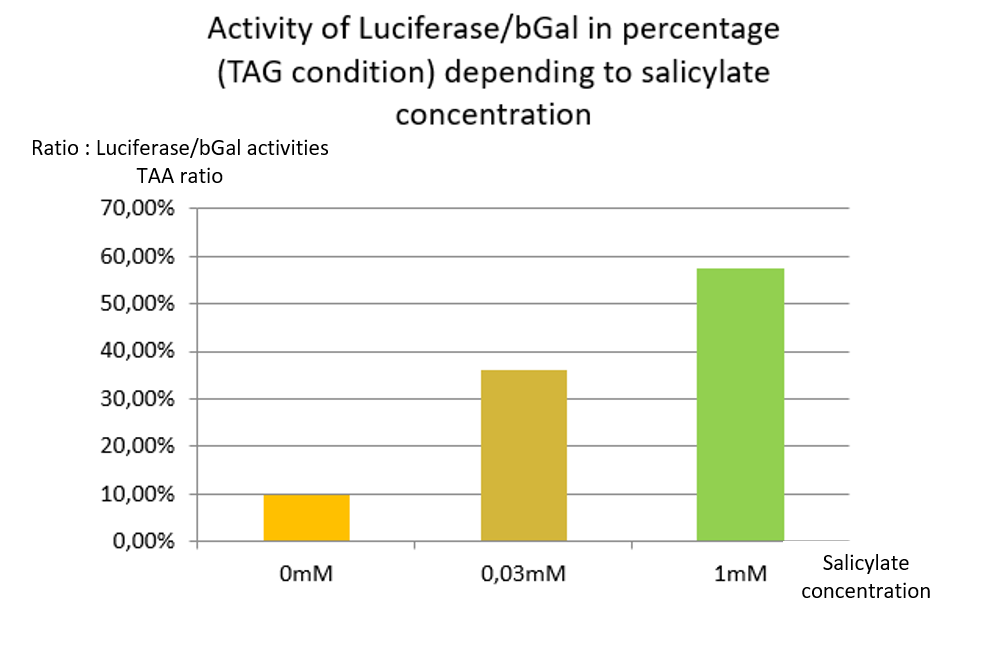
In TAG condition we can see an increase of stop codon readthrough activity with the increase of the salicylate concentration [Fig. 4].
In comparison to the results obtained with the TAA construction, the readthrough level increases similarly to the concentration of salicylate.
This indicates that TAG stop codon is efficiently readthrough in presence of supD tRNA, allowing the production of a significant amount of luciferase.
In conclusion, the Psal promoter is fully inducible by salicylate and the suppressor tRNA is functional to suppress the TAG codon. These experiments demonstrate that the BBa_K1372001 biobrick is fully functional.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 786
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 77
Illegal NgoMIV site found at 618
Illegal AgeI site found at 1309 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1312
Made by [http://2014.igem.org/Team:Paris_Saclay iGEM Paris Saclay]
| None |