Part:BBa_K1355003
Mercury ions accumulator device
We designed a biobrick device to bind and inactivate Hg. It is composed by two parts: Essential biobrick (BBa_K1355001) attached to the MBP biobrick (BBa_K346004) that was synthesized by iGEM Peking 2010. As described by the team, this MBP design was based on the tandem of two metal binding domains to make a high performance and less energy consuming metal binding peptide. The idea came from the high similarity of the C-terminal metal binding domain with MerR family TFs, which indicates a similar metal recognition mechanism and metal-protein complex structure.
The Mercury ions accumulator device biobrick (BBa_K1355003) has dual function: A) In reverse: transcription of MerR regulator protein; and B) In forward: transcription of MerP - MerT - Metal Binding Peptide proteins, as represented below:
In absence of mercury, MerR forms a MerR-promoter-operator complex, preventing RNA polymerase to recognize the promoter, consequently, messengers RNA for MerPT and MBP will not be transcript. In presence of Hg2+, MerR protein binds to this element and dissociates from the promoter-operator complex, allowing MerPT and MBP expression, as represented below:
When MerT, MerP and Metal Binding Protein (MBP) proteins are synthesized, the bioaccumulation will start! MerP and MerT protein are responsible for mercury’s transport of mercury from the periplasm to cytoplasm, leading it to MBP! The MBP binds to the mercury in the cytoplasm, inactivating it.
Mercury Bacter Hg bioaccumulator (DH5-alpha transformed with BBa_K1355003)
===Experiments and Results===
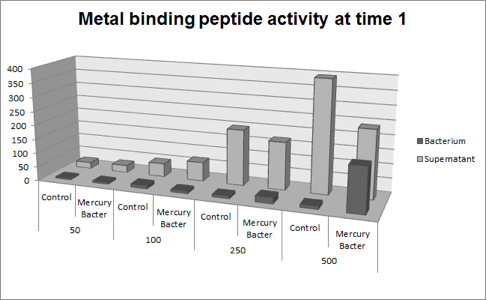
The experiment for Hg bioaccumulation was made according to the protocol “Quantification of Mercury bio accumulated by metal binding peptide (MBP) in recombinant DH5-alpha in different Hg concentrations”. DH5-alpha transformed with BBa_K1355003 was inoculated in LM (LB with low concentration of NaCl) liquid medium with chloramphenicol and grew until the optical density was 0.4 to 0.6abs (measured on spectrophotometer at 600 nm wavelength). After cell growth, an aliquot of 400μl was taken and distributed in 4 eppendorf tubes (1.5ml) and then added mercury chloride in order to achieve the concentrations: 50 ppb, 100 ppb, 250 ppb and 500 ppb. The samples were incubated at 37°C on shaker. We collected each eppendorf tube at time 1 (01:30 hours of incubation) and time 2 (03:00 hours of incubation). After the designated time, both were centrifuged at 12000g for 3 minutes and the supernatant recovered (LM medium). We washed the pellet with TN Buffer (Nacl 0.15M + Tris HCl 10mM) and then re-suspended with 400μl of the same buffer. To measure bio accumulated Hg, we need to quantify the Hg inside and outside of bacterium after the incubation/exposure time. So we collected and measured the amount of Hg in LM medium supernatant recovered and bacterium re-suspended in TN Buffer. For this we used the equipment Direct Mercury Analyzer (DMA-80). As a control to normal Hg bio accumulated in bacteria, we used DH5-alpha transformed with BBa_K1355002 (Hg bio detector device) which does not present the metal binding peptide. We also measured the Optical Density of each sample. The graph represented on Figure 1 shows the amount of Hg in supernatant (LM medium recovered) and in bacterium (DH5-alpha transformed with BBa_K1355003) at the time 1 (01:30 hours of incubation);
Figure 1: Metal binding peptide activity after 01:30 hours of incubation in five given concentrations of mercury chloride: 50 ppb, 100 ppb, 250 ppb e 500 ppb
It can be observed that the amount of Hg in the Mercury Bacter bioaccumulator increases according to the raise of Hg concentration. The amount of Hg increased 22 times comparing the 50 ppb sample with 500 ppb sample. In the 500 ppb Hg concentration the control bacterium just accumulated 2 per cent of total Hg amount. On the contrary, Mercury Bacter accumulated 40 per cent of total Hg amount in just 01:30 hours of incubation!!! The data keeps raising on the time 2! Check it out!
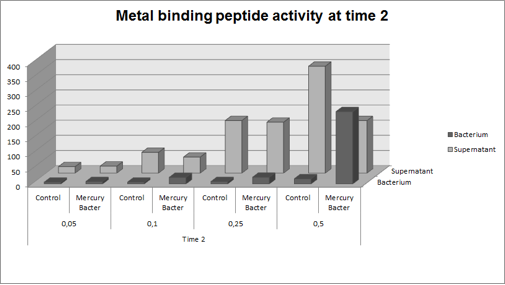
The graph represented on Figure 2 shows the amount of Hg in supernatant (LM medium recovered) and in bacterium (DH5-alpha transformed with BBa_K1355003) at the time 2 (03:00 hours of incubation);
Figure 2: Metal binding peptide activity after 03:00 hours of incubation in five given concentrations of mercury chloride: 50 ppb, 100 ppb, 250 ppb e 500 ppb.
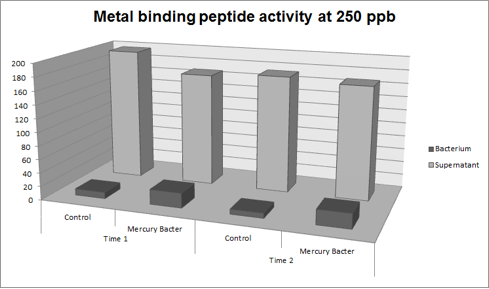
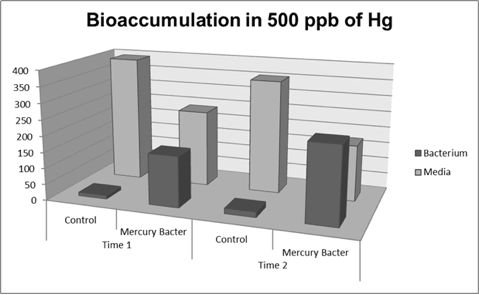
It can be observed that the amount of Hg in Mercury Bacter bioaccumulator, increases according to the time of incubation. The amount of Hg increased 30 times comparing the 50 ppb sample with 500 ppb sample. In the 500 ppb Hg concentration the control bacterium just accumulated 4% of total Hg amount. Instead, Mercury Bacter accumulated 58% of total Hg amount in just 03:00 hours of incubation!!! The data can be analyzed individually in each concentrations samples as shown in graphs represented in the figures below:
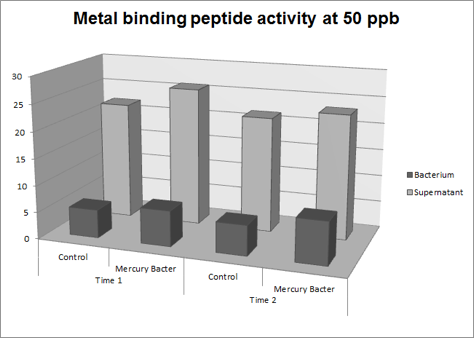
Figure 3: Metal binding peptide activity at 50 ppb in time 1 and 2;
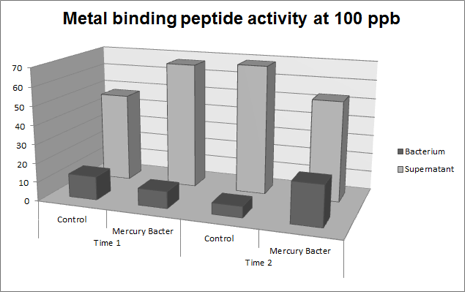
Figure 4: Metal binding peptide activity at 100 ppb in time 1 and 2;
Figure 5: Metal binding peptide activity at 250 ppb in time 1 and 2;
Figure 6: Metal binding peptide activity at 500 ppb in time 1 and 2
References
Gupta, R. K., Dobritsa, S. V., Stiles, C. A., Essington, M. E., Liu, Z., Chen, C. H., ... & Mullin, B. C. (2002). Metallohistins: A new class of plant metal-binding proteins. Journal of protein chemistry, 21(8), 529-536.
Mejáre, M., & Bülow, L. (2001). Metal-binding proteins and peptides in bioremediation and phytoremediation of heavy metals. TRENDS in Biotechnology, 19(2), 67-73.
Deng, X., & Wilson, D. (2001). Bioaccumulation of mercury from wastewater by genetically engineered Escherichia coli. Applied microbiology and biotechnology, 56(1-2), 276-279.
Licini, G., & Scrimin, P. (2003). Metal‐Ion‐Binding Peptides: From Catalysis to Protein Tagging. Angewandte Chemie International Edition, 42(38), 4572-4575.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 988
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 586
Illegal NgoMIV site found at 1160 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 579
| None |