Part:BBa_K1334002
HxlR+P-formaldehyde
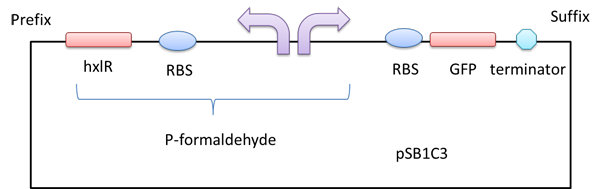
A right promoter which can sense formaldehyde.There is a left promoter which can erpress the HxlR protein.The HxlR protein can enhance the function of the P-formaldehyde.
Usage and Biology
A gfp reporter gene was added downstream so that we can know whether the P-formaldehyde works or not in the presence of formaldehyde by detecting the relative fluorescence intensity.
Figure 1 A GFP reporter is added downstream of the formaldehyde promoter.
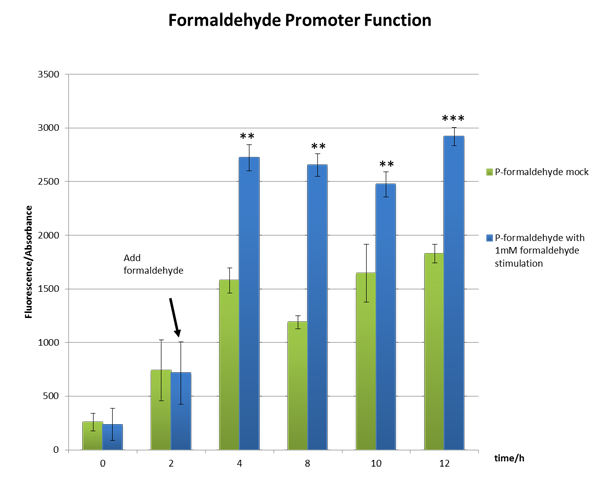
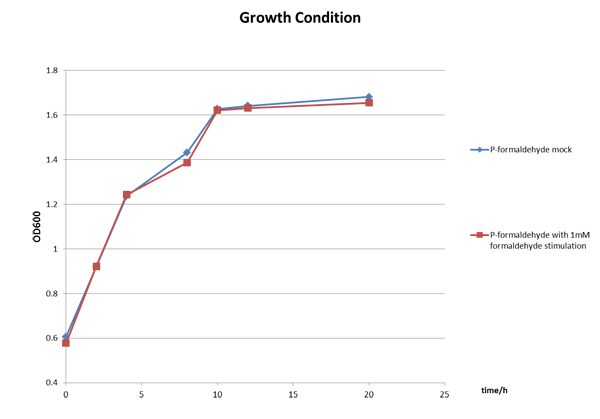
We incubated E.coli strain DH5a with the above plasmid (P-formaldehyde plus GFP)and stimulated the experimental group with 1mM formaldehyde when the OD600 value reached approximately one, which means E.coli comes into mid-log phase. And then we collected bacteria at certain time to measure the fluorescence and the absorbance.
From the Figure 2, we can see significant increase in relative fluorescence intensity after formaldehyde stimulation, which shows that our coloration system works well.
The Growth Condition curve in Figure 3 shows that the difference of the fluorescence/absorbance is not generated by the absorbance difference.
Figure 2 The function test of the formaldehyde promoter which shows our part BBa_K1334002 works well。
Figure 3 The growth condition of E.coli strain DH5a after formaldehyde stimulation.
Characterization from JNFLS 2019
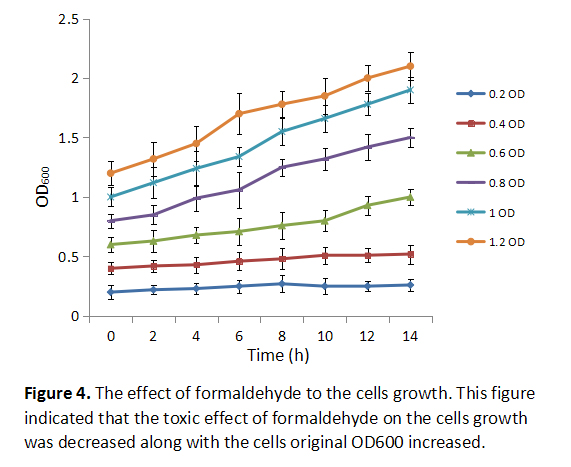
1.The effect of formaldehyde to the cells growth
Formaldehyde is toxic to the cells as we know, inhibiting cells growth if it is added early to the culture medium. In order to know at which OD600 the formaldehyde should be added into the culture medium, we added the 0.5mM formaldehyde to cells at different original OD600, then we collected different group bacteria at certain time to measure cells’ DO600 absorbance (Figure 4). The result showed that the toxic effect of formaldehyde on the cells growth was decreased with the cells original OD600 increased.
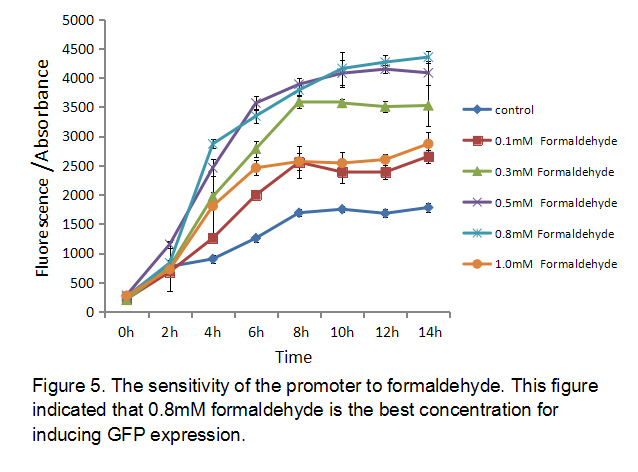
2.The sensitivity of the promoter to formaldehyde
Different concentration of formaldehyde has different ability to induce expression. In order to find the best concentration of formaldehyde to induce this promoter, we measured the sensitivity of the promoter to formaldehyde using BL21 as chassis. We cultured E.coli BL21 transformed with the plasmid (P-formaldehyde plus GFP)and added different concentration formaldehyde to different groups at OD600 = 1. Then we collected different group bacteria at certain time to measure the fluorescence and the absorbance (Figure 5). The result showed that 0.8 mM formaldehyde is the best concentration for inducing GFP expression.
Improve From XMU-China 2020
Group:iGEM Team XMU-China 2020
Author: Shuling Xiao
The formaldehyde promoter is one of the parts in our kill switch system. However, during our experiments, we found that the sensitivity of the current formaldehyde promoter is not good enough. Therefore, we are trying to find a way to improve the promoter.
Design
The formaldehyde promoter BBa_K1334002 contains hxlR coding sequence, a weak promoter, the formaldehyde promoter and two hxlR protein binding sites. Therefore, we figured out three ways below to improve its function.
Firstly, we changed the weak promoter to J23100, which is considered as a strong promoter in the Anderson family, and built BBa_K3332042.
Secondly, we added two more hxlR binding sites, creating BBa_K3332043.
Thirdly, we combined those two methods mentioned above and built BBa_K3332044, whose promoter has been changed and the binding sites are added.

- Fig 1. Gene circuit of formaldehyde promoter(BBa_K1334002).
Result
After designing different improvement of Formaldehyde promoters, we were verified together with the original formaldehyde promoter.

- Fig 2. Different improvement of Formaldehyde promoters
Three types of promoter improvements were designed as above. Firstly we’ve characterized the strength of J23100 and weak promoter by measuring the fluorescence/OD600 of J23100-B0034-eCFP and weak promoter-B0034-eCFP, promoter J23100 is far more stronger than weak promoter.

- Fig 3. Comparison of the strength of J23100 promoter and weak promoter by measuring the fluorescence/OD600
As the figure shown above, J23100 promoter can be used to enhance the strength of formaldehyde promoter as a strong promoter. However, the result of J23100 alternation was not significantly stronger than that compared to the original pHCHO. Therefore, we came out the other two improved version of pHCHO by adding extra binding site as well as both extra binding site and J23100, as the figure shown below.

- Table 1. Three version of modified pHCHO.
Then, the three version of promoters were jointed with fluorescence protein ECFP, and we detected the fluorescence of ECFP as an indication of promoter strength after formaldehyde induction, which activated the promoters and downstream gene expression.

- Fig 4. Fluorescence intensity/OD600 after inducing with 0.8 mM HCHO in 18 hours. Data are collected and analyzed according to iGEM standard data analysis.
From this figure, we can see that the fluorescence/OD600 of the induction group increased, but the improved promoters(modified-1~3) are more obvious than the registry formaldehyde promoter (BBa_K1334002).
Among the three modified promoters, the third modified version appeared to be the strongest one, showed the highest ECFP fluorescence intensity. That is to say the improved Formaldehyde promoters has a higher responsiveness to HCHO, which means that our three kinds of improvements work indeed.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |