Part:BBa_K1223006:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K1223006
User Reviews
UNIQ5963012ccf6d15dc-partinfo-00000000-QINU
|
No review score entered. franquero |
Characterization of BBa_K1223006 by 2019 MADRID_UCM teamCharacterization of this part has been conducted by building a transcriptional unit BBa_K3122004 and performing assays with nickel beads in order to check the interaction between them and its force. This transcriptional unit was assembled in pARK1 alpha plasmid including the following parts:
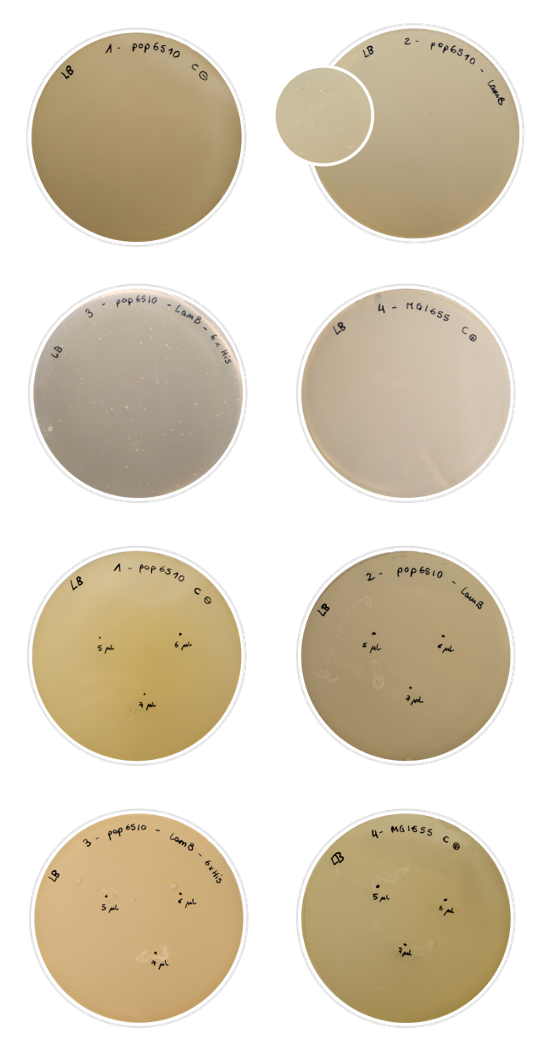
BackgroundThis part was used as a cell purification system for the SELEX procedure. Histidines may bind to nickel magnetic beads, allowing the separation and retention of the cells under a magnetic field. We decided to express a six-unit histidine tag in our LamB display system, in the permissive loop of LamB . The peculiarity of this tag is that histidines bind strongly to nickel beads subjected to a magnetic field. Since histidine is a polar amino acid, negatively charged (due to its multiple amine groups) in a wide range of pH, it can easily interact with ions of metallic particles (Thermofisher, 2019). Using this approach, we suggest that by expressing enough histidine tags in the membrane of our target cells we would be able to induce conjugation between magnetic beads and cells when incubated together in the magnetic module. For further information, visit our Robo-SELEX page . Besides, by using this promoter of medium strength, we ensure the expression of our protein without overburdening the cell membrane, and therefore, withput killing the cell. ExperimentsFor this characterization, we chose to perform a rather simple test that nonetheless gives conclusive results: the Lambda phage assay. The Lambda phage is a bacteriophage which infects the cell using the LamB protein. We used pop6510 cell line, which does not express LamB. Therefore, if there is no expression of our constructs, the phage cannot infect the cell and there will not be any lysis. On the other hand, if the Lamb display system is expressed properly, the phage will be able to infect the cell, and lysis spots shall appear in the Petri dish. In this experiment, we characterized the expression in the outer membrane of the LamB protein both with the 6xHis tag and without. We aimed to tell if we had achieved a correct expression of the protein, and if the addition of the His-tag into the permissive loop would compromise this expression. To prepare the test, we first dabbed the white colonies and made a liquid inoculum. The grown inoculum was treated as explained in the Fague lysis protocol, and then we conducted two different assays:
We used regular MG1655 E. coli cells (expressing LamB normally) as positive control, and our pop6510 cells as negative control. The results obtained were positive:
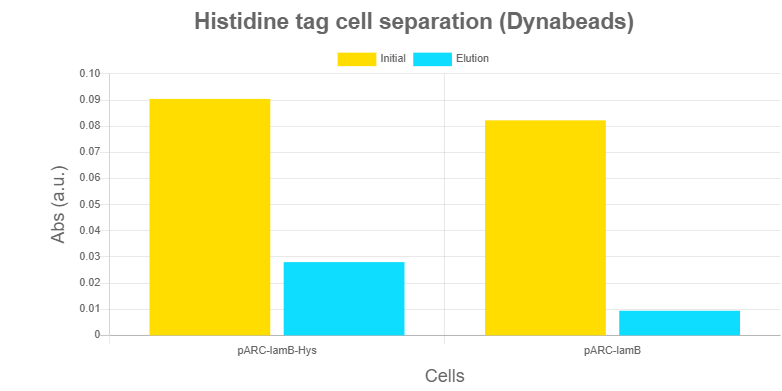
Characterization of Histidine-based separationHistidines are used as base of the separation system of our Robo-SELEX protocol. Several alternative strategies were designed, all based on the same concept: the LamB - 6xHis tag functions as an anchoring system, so the cell is retained while aptamers are eluted.
Results
ReferencesJ. Kim, C. Valencia, R. Liu and W. Lin, "Highly-Efficient Purification of Native Polyhistidine-Tagged Proteins by Multivalent NTA-Modified Magnetic Nanoparticles", Bioconjugate Chemistry, vol. 18, no. 2, pp. 333-341, 2007. Available:10.1021/bc060195l. |
UNIQ5963012ccf6d15dc-partinfo-00000009-QINU
UNIQ5963012ccf6d15dc-partinfo-0000000A-QINU
|
No review score entered. franquero |
Characterization of BBa_K1223006 by 2022 Thailand_RIS teamBackgroundSimilarly to what the 2019 Madrid team proposed, this part holds the potential to be used in other ways of separation. For our experiment, the Histidine tag was used as a part of purification through His column. Since Histidine contains polar properties, metal ions such as Ni2+ or Cu2+, will attract the histidine and allow for bonds to form. This attraction will form strong bonds that will cause the His-tags to form bonds with the linings on the side of the column, allowing for the washing of surplus proteins, without losing our target protein. ExperimentsFor our experiment, we had to undergo sonication to remove our protein from the host cell itself. However, when sonication occurs, even though the host cell is destroyed and proteins are free, all of the proteins are merged together. Thus, we had to separate our target protein from the rest of the proteins that kept the cells functioning. To accomplish this, we had to depend upon the His-tags. Within the amino acids of our peptide sequence Histidine which contains polar properties. With this, we decided to use His-columns, columns that are lined on the edges with metal ions, to separate our target protein from the rest. The histidine within our protein sequence will be attracted to the metal ions on the sides of the column due to the bonds that will be formed between the Histidine and metal ions. Since the other protein doesn’t contain Histidine in its sequences, no bonding will occur which will cause it to continually float in the solution mixture, separating it from our target protein. As we wash away the solution mixture, none of our target protein will be removed since it's still bonded to the sides. Afterwards, we wash the column with imidazole, eluting the protein. The imidazole is capable of eluting the protein since it competes with the histidine to bind with the metal ions lining the his columns. SDS-page after sonication of host cells. Targeted protein is shown as the highlighted bands. Other bands are different proteins that were present in host cells after Sonication. ResultsAfter eluting the column with imidazole, we ran the collected protein on gel electrophoresis. We determined if the protein is correct through the band sizes that are present. Our peptide, Bombolitin, requires band sizes of 125 bp. This shows that the protein that was collected was the correct one, with the base pairs of the bombolitin being between 100 and 200 base pairs. References“His-Tag Purification.” Bio, https://www.bio-rad.com/featured/en/his-tag-purification.html. |
UNIQ5963012ccf6d15dc-partinfo-0000000D-QINU