Part:BBa_K1100152
Csy4-16nt interaction estimation device
In this device, we tested the interaction between csy4 and its 16-nt cleavage site in vivo to be a supplementary experiment for the Rachel E. Huarwitz et al, and Lei Qi et al's previous work.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal prefix found in sequence at 67
Illegal SpeI site found at 885
Illegal PstI site found at 1775 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 67
Illegal SpeI site found at 885
Illegal PstI site found at 1775
Illegal NotI site found at 73 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 67
- 23INCOMPATIBLE WITH RFC[23]Illegal prefix found in sequence at 67
Illegal SpeI site found at 885
Illegal PstI site found at 1775 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found in sequence at 67
Illegal XbaI site found at 82
Illegal SpeI site found at 885
Illegal PstI site found at 1775
Illegal NgoMIV site found at 536 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 252
Background
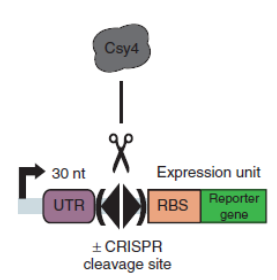
Csy4 is a member of CRISPR pathway discovered in Pseudomonas aeruginosa. It is a single endoRNase that recognizes and cleaves a 28-nucleotide repetitive sequence and produces stable transcripts with a 5’hydroxylgroup, which can eliminate unwanted interactions between 5’UTRs and translational elements such as RBSs units to standardize the expression of the elements. ( Lei Qi et al, 2012).The cleavage site can be curtailed to 16nt proved by in vitro data (Rachel E. Huarwitz et al, 2010)
Figure. 1. The CRISPR RNA-processing system facilitate engineering of standard genetic elements in various contexts( Lei Qi et al, 2012).
Design
We introduced the csy4 platform and its cleavage platform onto one plasmid as followed: pSB1C3-BBa_R0040-16nt-BBa_B0034-Csy4-BBa_B0015-BBa_R0063-16nt-BBa_B0034-sfGFP, in order to let the csy4 express to recognize and cleave the 16-nucleotide sequence.
Reference:
Haurwitz R E, Jinek M, Wiedenheft B, et al. Sequence-and structure-specific RNA processing by a CRISPR endonuclease[J]. Science, 2010, 329(5997): 1355-1358. Qi L, Haurwitz R E, Shao W, et al. RNA processing enables predictable programming of gene expression[J]. Nature biotechnology, 2012, 30(10): 1002-1006. Cambray G, Guimaraes J C, Mutalik V K, et al. Measurement and modeling of intrinsic transcription terminators[J]. Nucleic acids research, 2013, 41(9): 5139-5148.
| abs | B0034 |
| biology | |
| chassis | E.coli |
| control | aTc, tetracyline |
| direction | Forward |
| efficiency | 1 |
| emission | 510nm |
| excitation | 485nm |
| negative_regulators | 1 |
| o_h | |
| o_l | |
| positive_regulators | |
| protein | sfGFP |