Part:BBa_K1075044
Promoter(const.)-RBS34-YF1-FixJ-FixK-LambdaC-pC (pDawn)
pDawn can be used to induce transcription of a downstream gene with blue light.
The plasmid pDawn was designed by Ohlendorf et al. in 2012 together with its counter plasmid pDusk. Both plasmids are single plasmid systems, which allow the activation (pDawn) or repression (pDusk) of gene expression by blue light. They are easy to implement in the laboratory and lead to up to 460-fold activity change upon ilumination.
Usage and Biology
Ohlendorf et al. (2012) had previously designed the histidine kinase YF1, which phosphorylates FixJ in the absence of blue light. The phosphorylated FixJ drives robust gene expression from the FixK2 promotor as shown in figure 1 for both pDawn and the pDusk. Upon light absorption, net kinase activity of YF1 and consequently gene expression under control of the FixK2 promotor is greatly reduced. pDusk an pDawn enable light-repressed or light-induced gene expression in Escherichia coli and are easy to implement in the laboratory. Therefore, all components of the YF1/FixJ TCS were assembled on only one medium-copy plasmid in which YF1 and FixJ are constitutively expressed by the Lacq promotor. Target genes can be introduced via the multiple-cloning site (MCS) under the control of the pFixK2 promotor, allowing light-repressed gene expression. To obtain pDawn the light effect is inverted by the introduction of a gene-inversion cassette into pDusk. This gene-inversion cassette is based on the λ phage repressor cI and the λ phage promotor pR.
Ohlendorf et al. (2012) analyzed the system by the insertion of the red-fluorescent reporter protein DsRed Express2. That way they were able to measure up to 460-fold induction upon ilumination. However, they also found that their plasmid, which acts on gene expression level, only allows light-induced perturbations on the timescale of hours.
Further Characterization I
In addition to the literature data, further testing was conducted in 2017 by Cornell iGEM. By growing E. coli under different intensities of visible light, we can compare the relative expression levels of a FLAG-tagged control protein, superoxide dismutase, under optogenetic control.
We first inserted superoxide dismutase into the pDusk circuit. We then proceeded to incorporate a cI repressor (BBa_K2296045) and a pR promoter into the pDusk system followed by insertion of superoxide dismutase to create a light-inducible pDawn circuit.
We assessed the transcriptional activity of pDawn and pDusk in response to three different levels of light intensity from a standard laboratory fluorescent light:

Our initial characterization revealed a dose-dependence of pDusk. However, pDawn did not display a dose-dependent response in response to stimulation from fluorescent light. We suspect this to be the result of low light intensity.
Our subsequent characterization uses a significantly greater intensity of LED light:

Our results suggest a light-intensity dependence of pDusk and pDawn. Our results also show that our cI repressor biobrick successfully converted the light-repressible pDusk into a light-inducible pDawn.
In addition to dose-dependent characterization, we also conducted time-dependence characterization of pDawn following stimulation by 470 nm LED light (LTL3H3TBPADS1) with the following emission spectrum [2]:

Our initial characterization was conducted with 1 second pulses and revealed a significant lag between initial exposure and translational response:

For our subsequent characterization, we increased the intensity and pulse duration of the LED to parse out more subtle effects at earlier time points:

Our results suggest a light intensity and time-dependence of the pDawn and pDusk optogenetic circuit.
Further Characterization II
ShanghaiTech iGEM(https://2019.igem.org/Team:ShanghaiTech_China) characterized the efficiency of pDawn system with mEGFP and improved it.
Results were analyzed in both visualized micrographs and normalized line graphs, which showed the qualitative and quantitative mEGFP expression level respectively.
Micrographs are listed as follows:
pDawn with T7 promoter is abbreviated as T7c below. pDawn with tac promoter is abbreviated as tac below. pDawn with modified tac promoter is abbreviated as mtac below. pDawn with YF2 is abbreviated as YF2 below.
[Go for the protocol of characterization:https://2019.igem.org/Team:ShanghaiTech_China/LightControl]
Result: As the illumination duration increases, the bacteria densities also increased until it became stable after 30 hours. Substantially, there were also big increases in mEGFP fluorescence per bacterium in light-on groups over light-off groups, suggesting the light induction was successful.
Result: The graph shows that mEGFP expression was sharply increased after 6 hours when treated with light, and reached the maximum expression level after 24 hours. Although mEGFP expression in the light-on groups were much higher than that in light-off groups, there was significant leakiness expression in darkness. To overcome this deficiency, several modifications were applied to the plasmid, which will be elucidated in the next part.
To see more changes on pDawn, go to the websites below.
pDawn with T7 promoter: https://parts.igem.org/Part:BBa_K3278001
pDawn with mtac promoter: https://parts.igem.org/Part:BBa_K3278002 (better)
pdawn with tac promoter: https://parts.igem.org/Part:BBa_K3278005
pDawn with YF2: https://parts.igem.org/Part:BBa_K3278006 (better)
References
1 [http://www.sciencedirect.com/science/article/pii/S0022283612000113] Ohlendorf et al. (2009) From Dusk till Dawn One-Plasmid Systems for Light-Regulated Gene Expression.
2 Lite-On Technology Corporation. Information Data Sheet for LED Lamp LTL3H3TBPADS1-132A. Retrieved from http://www.mouser.com/ds/2/239/Lite-On_LTL3H3TBPADS1-132A-Ver.A-341105.pdf
2022 SZPT-China
Characterization
The leakage level of the original pDawn system in which the repressor cI contains a protein degradation tag LVA and that of the modified pDawn system in which the LVA tag of cI is removed are compared in a kill switch using lysis cassette LKD.
The plasmid containing the blue light induced kill switch was transformed into E. coli TOP10. Sixteen colonies were picked and cultured on two new LB plates at 37 °C for 16 hours. One plate was placed in the dark and the other was placed under blue light. The number of the candidates that survived the dark but failed to grow under blue light was counted. For the E. coli strain containing pDawn(cI-LVA)-RBS070-LKD, the number is 5. For pDawn(cI)-RBS070-LKD, the number is 10. The leakage expression of the lysis cassette LKD could be associated with this discrepancy in functionality. The new pDawn in which the LVA tag of cI is deleted improves the effectiveness of the blue light responsive lysis system.

(a2) Colony growth of E. coli TOP10 containing the plasmid of pDawn(cI-LVA)-RBS070-LKD under blue light
(b1) Colony growth of E. coli TOP10 containing the plasmid of pDawn(cI)-RBS070-LKD in the dark
(b2) Colony growth of E. coli TOP10 containing the plasmid of pDawn(cI)-RBS070-LKD under blue light
From the above two groups of successful recombinant candidates (red squares in Figure 1), one colony was selected and cultured on a LB plate respectively. For each strain, sixteen colonies were picked and cultured on two new LB plates at 37 °C in the dark. The growth condition of the bacteria on each plate was observed after 12-16 hours. We found that for the strain containing pDawn(cI-LVA)-RBS070-LKD, a significant proportion of the recombinant colonies did not grow very well in the dark, indicating a leak expression of the lysis genes. Nevertheless, for the strain containing pDawn(cI)-RBS070-LKD, all the colonies grow normally in the dark. Therefore, the new pDawn in which the LVA tag of cI is deleted reduces the fitness cost of the blue light responsive lysis system, while the system with the original pDawn confers a significant growth disadvantage.
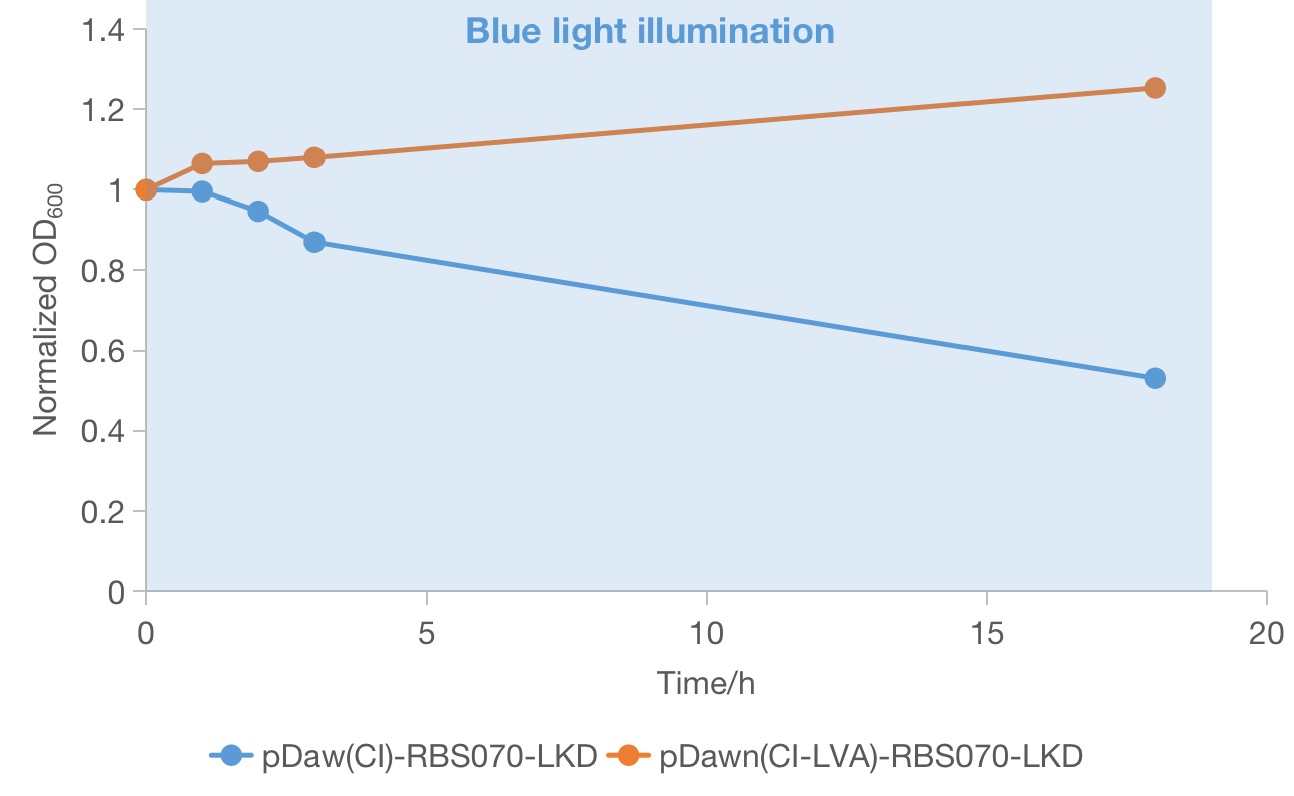
From the above two groups of successful recombinant candidates (red squares in Figure 1), one colony was picked and grow in LB medium in the dark until ~OD600 0.6 was reached to track the growth condition under blue light. Then the OD600 values of the two selected strains under blue light was measured to compare the effectiveness of the two systems in rapidly growing cells. We found that both of the strain continued to grow during the first few hours of blue light illumination. However, only the strain containing the plasmid of pDawn(cI)-RBS070-LKD showed a significant decrease in OD600 value, indicating maintenance of lethality. And the OD600 value of the strain containing the plasmid of pDawn(cI)-RBS070-LKD increases a little bit, implying that loss-of-function mutations could occurred in these genes or in the host genome and that the favorable mutant overtakes the parental population. Therefore, the new pDawn in which the LVA tag of cI is deleted increases the evolutionary stability of the blue light sensitive kill switch by negating the deleterious evolutionary pressure of leaky toxin expression.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2171
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 63
Illegal NgoMIV site found at 195
Illegal NgoMIV site found at 289
Illegal NgoMIV site found at 582
Illegal NgoMIV site found at 1076
Illegal NgoMIV site found at 1094
Illegal NgoMIV site found at 1184
Illegal AgeI site found at 414
Illegal AgeI site found at 1542 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1643
Illegal BsaI.rc site found at 525
| None |