Part:BBa_J23113:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_J23113
Evaluation of Anderson promoter J23113 in B. subtilis by iGEM-Team LMU-Munich 2012
This Anderson promoter was evaluated without fused RFP with the lux operon as a reporter in B. subtilis. See the new BioBrick BBa_K823010 without RFP and have a look at the [http://2012.igem.org/Team:LMU-Munich/Data/Anderson Data] from the evaluation in B. subtilis.
User Reviews
UNIQ81a1d139b1fced10-partinfo-00000000-QINU UNIQ81a1d139b1fced10-partinfo-00000001-QINU
|
•••••
for iGEM-Team Goettingen 2012 |
Characterization experiment by qrtPCR on BBa_J23100, BBa_J23104, BBa_J23105, BBa_J23106, BBa_J23109, BBa_J23112, BBa_J23113, BBa_J23114 by iGEM Team Göttingen (by C. Krüger and J. Kampf)DescriptionWe used quantitative real-time PCR as a powerful tool for quantification of gene expression. We used this method to examine the expression rate of the Tar receptor gene under control of promoters from the Anderson family of the parts registry. The BioBricks (K777001-K777008) we used for this experiment can be found here. The reported activities of these promoters are given as the relative fluorescence of these plasmids in strain TG1 [1]. Promoter constructs were cloned into the vector pSB1C3 and expressed in E.coli BL21DE3 grown in LB-media (lysogeny broth). The measurements were performed for each construct and reference as a triplet. Additionally, we included H2O as negative control to predict possible contamination. For the evaluation of our results, the 2–ΔΔCT (Livak) method was applied. We used the weakest promoter with the lowest expression rate as calibrator for the calculations and as reference the housekeeping gene rrsD of E.coli.
You can find detailed information of the qrtPCR approach [http://2012.igem.org/Team:Goettingen/Project/Methods#-.3E_Experimental_design here].
Results & Discussion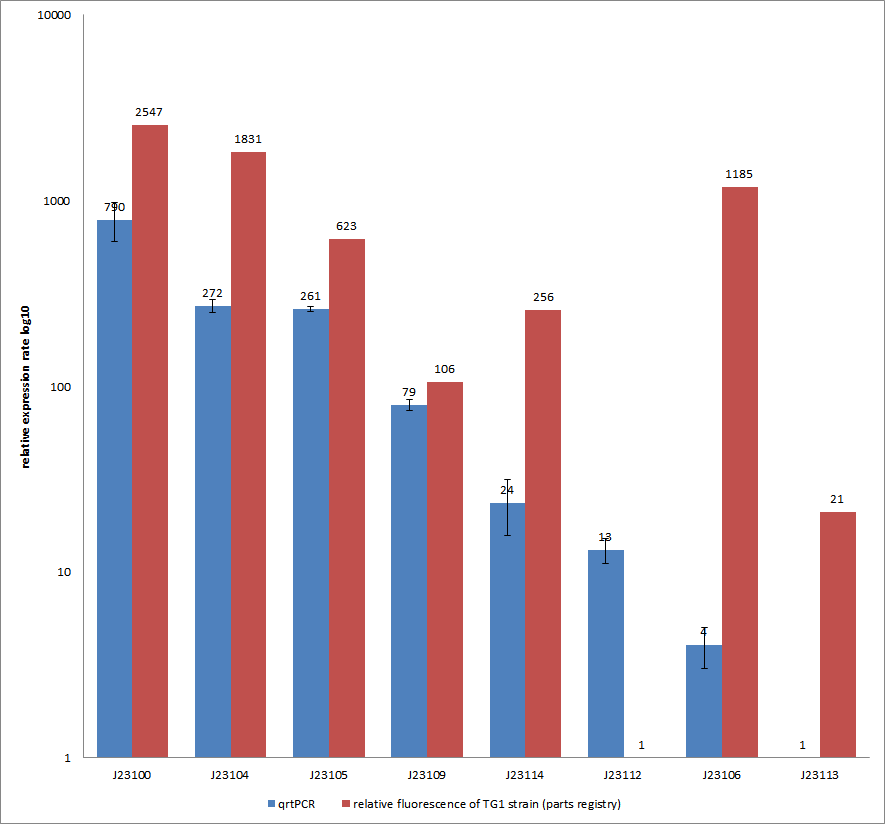 Comparison of relative expression rates of constitutive promoters by qrtPCR and relative fluorescence (see parts registry,Anderson family). The blue bar indicates the measured expression rates for our constructs (J23100, J23104, J23105, J23106, J23109, J23112, J23113, J23114) and the red ones those for the literature values represented in the “parts registry”. The measurements are illustrated in a logarithmic application. The standard variation was calculated for our measured values (black error bar). As mentioned before, both datasets were collected by methods which produce data at different points after the gene expression. Quantitative real time PCR measures the amount of expressed mRNA while relative fluorescence measurements quantify on protein level. In perspective of stability and half-life periods of mRNA and proteins or due to protein modification, it is comprehensible to obtain varying data-sets and expression rates. Another problem that occurred during our quantitative real-time measurements was the deviation in some of biological replicates. This problem was also observed in another group’s experiments ([http://www.jbioleng.org/content/3/1/4 Kelly et al., 2009]). They mentioned variations across experimental conditions in the absolute activity of the BioBricks. To reduce variation in promoter activity, they measured the activity of promoters relative to BBa_J23101. Furthermore, the iGEM team of Groningen which participated in 2009 also measured the relative fluorescence of TG1 strain with the promoters J23100, J23109 and J23106 via Relative Promoter Units (RPUs). Their values indicated the comparable tendency to our documented values |
|
•••••
University of Texas at Austin iGEM 2019 |
UT Austin iGEM 2019: Characterization of metabolic burden of the Anderson SeriesDescriptionThe 2019 UT Austin iGEM team transformed the Anderson Series promoters into our 'burden monitor' DH10B strain of E. coli, which contains a constitutive GFP cassette in the genome of the cell. GFP expression fluctuates depending on the number of ribosomes available. Using this strain, we characterized the relative burden (percent reduction in growth rate) of each Anderson Series part. Our results showed a range of growth rate reductions for each of these parts due to ribosomal reallocation from the genome of the host cell, towards the expression of RFP. Anderson Series parts with strong promoters are depicted with darker red colors and Anderson Series parts with weak promoters are depicted with lighter pink colors to show relative RFP expression. We saw a positive correlation between relative promoter strength and metabolic burden; parts with stronger promoters expressed less GFP and had a lower growth rate than parts with weaker promoters. The regression line for the graph below was constructed by measuring the burden of 5 parts that were created by the 2019 UT Austin iGEM team that each contained an Anderson Series promoter (BBa_J23104 or BBa_J23110), an RBS of varying strength, and a BFP reporter. For more information on characterization of these parts through the burden monitor, visit our team’s wiki page: [2]
Importance of Characterizing BurdenAlthough often we cannot avoid using a specific burdensome part, knowing in advance that it is burdensome, and that it has a high chance of mutating into a non-functional genetic device, can help with troubleshooting and coming up with alternatives. In the specific case of fluorescent protein-expressing devices, Fluorescence-activated cell sorting (FACS) can be used to filter out individual cells that meet a certain fluorescence threshold. This way, the cells expressing lower levels of the fluorescent protein are weeded out of the population. |

 1 Registry Star
1 Registry Star
