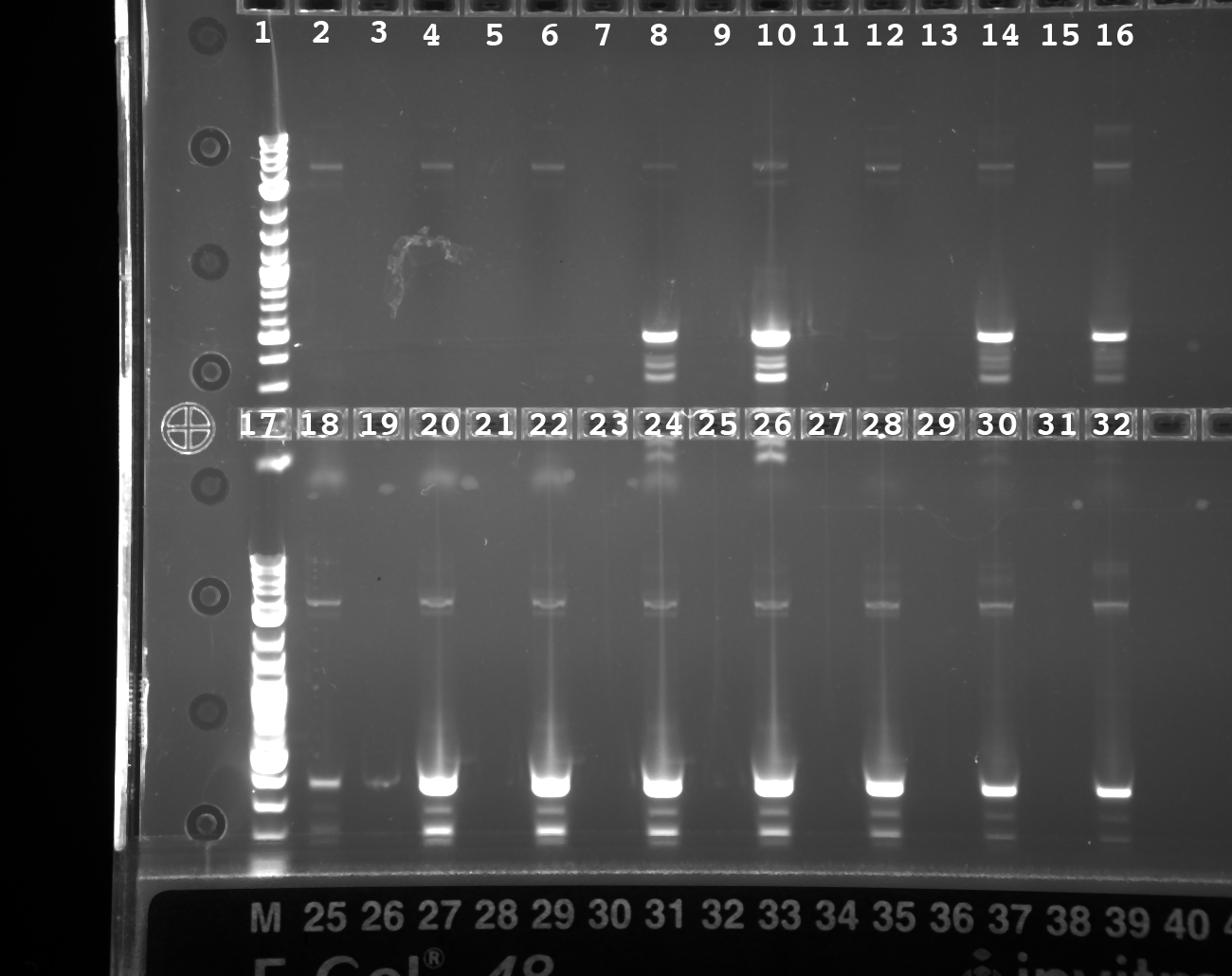Difference between revisions of "VR/VF2 Gradient PCR Experiment"
m (→Experiment) |
(→Results) |
||
| Line 28: | Line 28: | ||
== Results == | == Results == | ||
| − | [[Image:070817.png|thumb|right|Results of Gradient PCR]] | + | [[Image:070817.png|thumb|right|Results of Gradient PCR - S03582 is in the top row, I13033 is in the bottom row]] |
| − | + | As you can see on the image of the gel, the extra bands still appeared at the different annealing temperatures. Samples were loaded in every other well, in order of increasing annealing temperature. | |
Revision as of 17:29, 28 August 2007
I tried gradient PCR to see if a higher annealing temperature would prevent the incorrect primer binding.
Experiment
I prepared 8 reactions for both S03582 and I13033
PCR Reaction
- 9 μL PCR Supermix High Fidelity (Invitrogen)
- .25 μL of 40 μM VR
- .4 μL of 25 μM VF2
- I ran out of 40 μM VF2 so I used more of a less concentrated sample to make up the difference
- 1 μL template DNA (~10 ng/μL)
- Initial denature 95°C - 5 min
- 35 cycles
- 94°C - 30 sec
- Gradient from 54°C to 70°C - 30 sec
- 68°C - 36 sec
- Final extension 68° - 10 min
- 4°C forever
*The annealing temperature gradient ran from 54°C to 70°C, but due to the placement of 8-well strips samples were at the following annealing temperatures: 55.4°C, 56.7°C, 58.5°C, 60.9°C, 63.6°C, 65.8°C, 67.6°C, 68.7°C
I ran the results on a 1% Agarose gel for 30 minutes.
Results
As you can see on the image of the gel, the extra bands still appeared at the different annealing temperatures. Samples were loaded in every other well, in order of increasing annealing temperature.