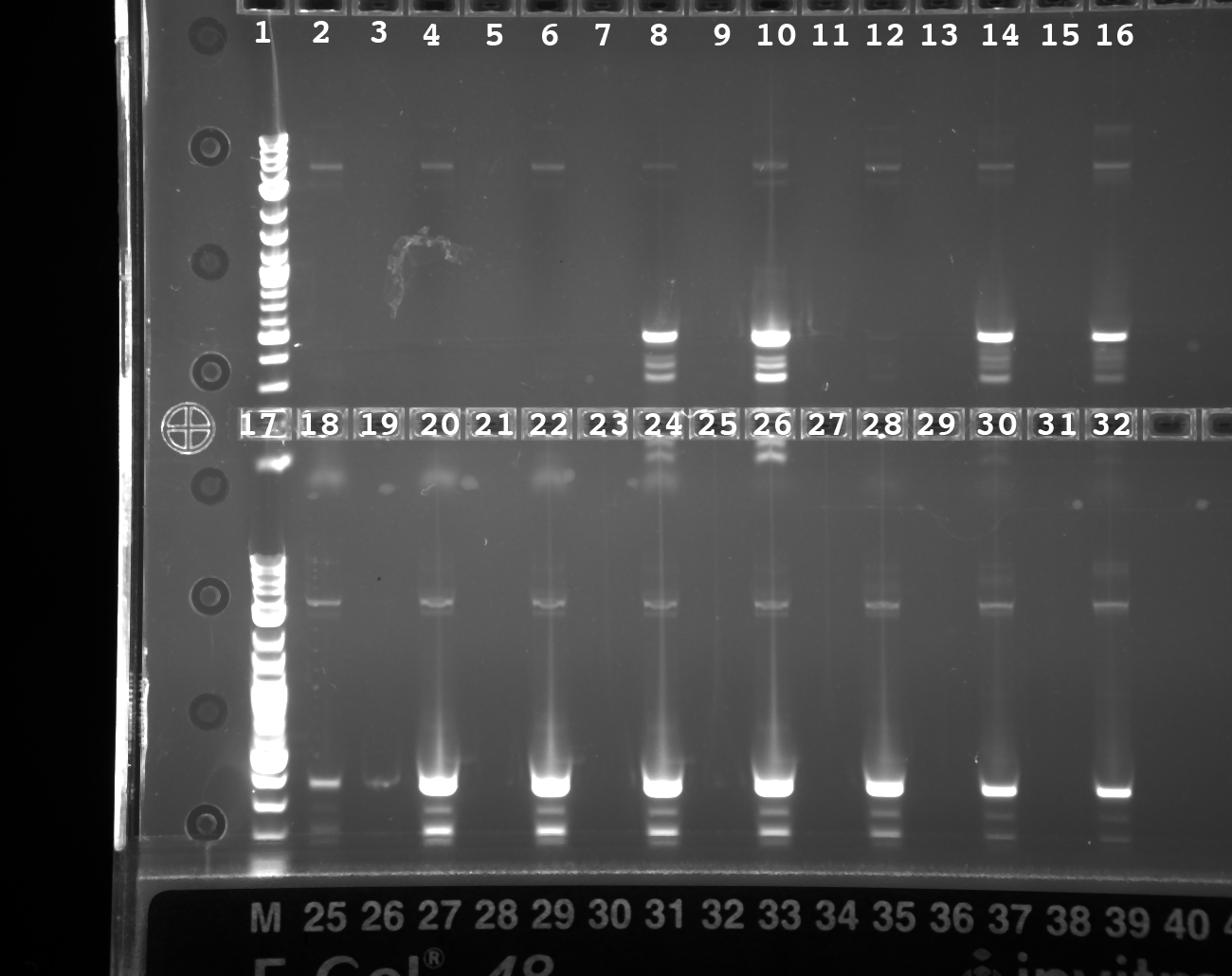Difference between revisions of "VR/VF2 Gradient PCR Experiment"
(→Results) |
m (→Experiment) |
||
| Line 6: | Line 6: | ||
'''PCR Reaction''' | '''PCR Reaction''' | ||
| − | *9 μ | + | *9 μL PCR Supermix High Fidelity (Invitrogen) |
*.25 μL of 40 μM VR | *.25 μL of 40 μM VR | ||
*.4 μL of 25 μM VF2 | *.4 μL of 25 μM VF2 | ||
Revision as of 17:15, 28 August 2007
I tried gradient PCR to see if a higher annealing temperature would prevent the incorrect primer binding.
Experiment
I prepared 8 reactions for both S03582 and I13033
PCR Reaction
- 9 μL PCR Supermix High Fidelity (Invitrogen)
- .25 μL of 40 μM VR
- .4 μL of 25 μM VF2
- I ran out of 40 μM VF2 so I used more of a less concentrated sample to make up the difference
- 1 μL template DNA (~10 ng/μL)
- Initial denature 95°C - 5 min
- 35 cycles
- 94°C - 30 sec
- Gradient from 54°C to 70°C - 30 sec
- 68°C - 36 sec
- Final extension 68° - 10 min
- 4°C forever
*The annealing temperature gradient ran from 54°C to 70°C, but due to the placement of 8-well strips samples were at the following annealing temperatures: 55.4°C, 56.7°C, 58.5°C, 60.9°C, 63.6°C, 65.8°C, 67.6°C, 68.7°C
I ran the results on a 1% Agarose gel for 30 minutes.
Results
The extra bands still appeared at the different annealing temperatures.