Difference between revisions of "Part:BBa K823039"
| Line 3: | Line 3: | ||
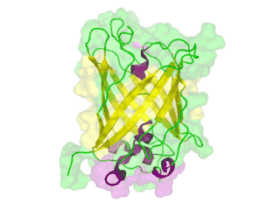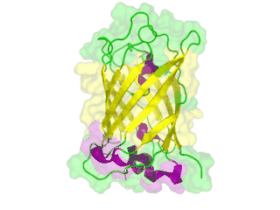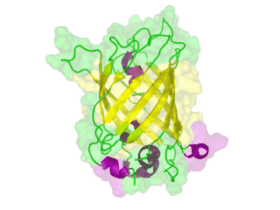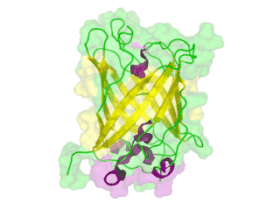
[[File:TUM13 Animation_GFP.gif|thumb|right|320px| Figure: Animated structure of the green fluorescent protein (GFP) based on the cristall structure [http://www.rcsb.org/pdb/explore.do?structureId=2WUR 2WUR] | [[File:TUM13 Animation_GFP.gif|thumb|right|320px| Figure: Animated structure of the green fluorescent protein (GFP) based on the cristall structure [http://www.rcsb.org/pdb/explore.do?structureId=2WUR 2WUR] | ||
with an identitiy of 98% with this BioBrick. Added by [http://2013.igem.org/Team:TU-Munich TUM 2013 Team]. ]] | with an identitiy of 98% with this BioBrick. Added by [http://2013.igem.org/Team:TU-Munich TUM 2013 Team]. ]] | ||
| − | This gfpmut1 gene was amplified from the ''B. subtilis'' vector pGFPamy. | + | This ''gfpmut1'' gene was amplified from the ''B. subtilis'' vector pGFPamy. |
| − | Big thanks to the TU-Munich TUM 2013 Team for the advice that this actually is gfpmut1 (not gfpmut3). | + | Big thanks to the TU-Munich TUM 2013 Team for the advice that this actually is ''gfpmut1'' (not ''gfpmut3''). |
It is constructed in the [[Help:Assembly_standard_25|Freiburg standard]] with RBS. | It is constructed in the [[Help:Assembly_standard_25|Freiburg standard]] with RBS. | ||
| Line 17: | Line 17: | ||
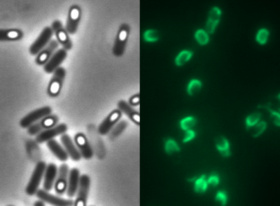
<p align="justify">[[Image:LMU Firstspore.jpg|200px|left]]We took a ''gfp'' derivate of the ''Bacillus subtilis'' plasmids pGFPamy and added the BioBrick compatiple pre- and suffix of the Freiburg standard (Assembly 25). This BioBrick was used to create our [http://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins '''Sporo'''beads]. It is part of the BioBrick [https://parts.igem.org/Part:BBa_K823049 K823049]</p> | <p align="justify">[[Image:LMU Firstspore.jpg|200px|left]]We took a ''gfp'' derivate of the ''Bacillus subtilis'' plasmids pGFPamy and added the BioBrick compatiple pre- and suffix of the Freiburg standard (Assembly 25). This BioBrick was used to create our [http://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins '''Sporo'''beads]. It is part of the BioBrick [https://parts.igem.org/Part:BBa_K823049 K823049]</p> | ||
| − | Find out more about our [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks ''<b>Bacillus'' B</b>io<b>B</b>rick <b>B</b>ox] and the created [http://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins '''Sporo'''beads]. | + | Find out more about our [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks ''<b>Bacillus'' B</b>io<b>B</b>rick <b>B</b>ox] and the created [http://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins '''Sporo'''beads]. This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The ''Bacillus'' BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with ''Bacillus subtilis''] by Radeck ''et al.''. |
<br> | <br> | ||
Latest revision as of 17:07, 3 February 2014
gfpmut1 (B. subtilis optimized + Freiburg standard)
This gfpmut1 gene was amplified from the B. subtilis vector pGFPamy.
Big thanks to the TU-Munich TUM 2013 Team for the advice that this actually is gfpmut1 (not gfpmut3).
It is constructed in the Freiburg standard with RBS.
Find out more about the design of our prefix with ribosome binding site.
prefix: GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC
suffix: ACCGGTTAATACTAGTAGCGGCCGCTGCAGT
We took a gfp derivate of the Bacillus subtilis plasmids pGFPamy and added the BioBrick compatiple pre- and suffix of the Freiburg standard (Assembly 25). This BioBrick was used to create our [http://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins Sporobeads]. It is part of the BioBrick K823049Find out more about our [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks Bacillus BioBrick Box] and the created [http://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins Sporobeads]. This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis] by Radeck et al..
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 197