Difference between revisions of "Part:BBa K4325010"
(→2022 SZPT-China) |
(→2022 SZPT-China) |
||
| Line 12: | Line 12: | ||
[[ File:Improvement3.jpeg|600px|thumb|center||Figure 1. Sequence alignment of pDawn(cI-LVA) before and after removing LVA tag]] | [[ File:Improvement3.jpeg|600px|thumb|center||Figure 1. Sequence alignment of pDawn(cI-LVA) before and after removing LVA tag]] | ||
<h3>Usage</h3> | <h3>Usage</h3> | ||
| − | <p>The plasmid containing the blue light induced kill switch was transformed into <i>E. coli</i> TOP10. Sixteen colonies were picked and | + | <p>The plasmid containing the blue light induced kill switch was transformed into <i>E. coli</i> TOP10. Sixteen colonies were picked and cultured on two new LB plates at 37 °C for 16 hours. One plate was placed in the dark and the other was placed under blue light. The number of the candidates that survived the dark but failed to grow under blue light but survived in the dark was counted. For the <i>E. coli</i> strain containing pDawn(cI-LVA)-RBS070-LKD-pSEVA331, the number is 5. For pDawn(cI)-RBS070-LKD-pSEVA331, the number is 10. The leakage expression of the lysis cassette LKD could be associated with this discrepancy in functionality. The new pDawn in which the LVA tag of cI is deleted improves the effectiveness of the blue light responsive lysis system.</p> |
[[File:Improvement4.jpeg|800px|thumb|center||Figure 2. (a1) Colony growth of <i>E. coli</i> TOP10 containing the plasmid of pDawn(cI-LVA)-RBS070-LKD-pSEVA331 in the dark | [[File:Improvement4.jpeg|800px|thumb|center||Figure 2. (a1) Colony growth of <i>E. coli</i> TOP10 containing the plasmid of pDawn(cI-LVA)-RBS070-LKD-pSEVA331 in the dark | ||
<p>(a2) Colony growth of <i>E. coli</i> TOP10 containing the plasmid of pDawn(cI-LVA)-RBS070-LKD-pSEVA331 under blue light </P> | <p>(a2) Colony growth of <i>E. coli</i> TOP10 containing the plasmid of pDawn(cI-LVA)-RBS070-LKD-pSEVA331 under blue light </P> | ||
Revision as of 12:03, 10 October 2022
pDawn without LVA tag
Description
Our reduce the leakage level of pDawn system by removing the degradation tag LVA in the original repressor cI .
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2171
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 63
Illegal NgoMIV site found at 195
Illegal NgoMIV site found at 289
Illegal NgoMIV site found at 582
Illegal NgoMIV site found at 1076
Illegal NgoMIV site found at 1094
Illegal NgoMIV site found at 1184
Illegal AgeI site found at 414
Illegal AgeI site found at 1542 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1643
Illegal BsaI.rc site found at 525
2022 SZPT-China
Biology
Our goal is to reduce the leakage level of pDawn system by removing the degradation tag LVA in the original repressor cI as increasing the concentration of cI could enhance the repression of pR promoter.
Usage
The plasmid containing the blue light induced kill switch was transformed into E. coli TOP10. Sixteen colonies were picked and cultured on two new LB plates at 37 °C for 16 hours. One plate was placed in the dark and the other was placed under blue light. The number of the candidates that survived the dark but failed to grow under blue light but survived in the dark was counted. For the E. coli strain containing pDawn(cI-LVA)-RBS070-LKD-pSEVA331, the number is 5. For pDawn(cI)-RBS070-LKD-pSEVA331, the number is 10. The leakage expression of the lysis cassette LKD could be associated with this discrepancy in functionality. The new pDawn in which the LVA tag of cI is deleted improves the effectiveness of the blue light responsive lysis system.

(a2) Colony growth of E. coli TOP10 containing the plasmid of pDawn(cI-LVA)-RBS070-LKD-pSEVA331 under blue light
(b1) Colony growth of E. coli TOP10 containing the plasmid of pDawn(cI)-RBS070-LKD-pSEVA331 in the dark
(b2) Colony growth of E. coli TOP10 containing the plasmid of pDawn(cI)-RBS070-LKD-pSEVA331 under blue light
From the above two groups of successful recombinant candidates (red squares in Figure 1), one colony was selected and streaked on a LB plate respectively. For each strain, sixteen colonies were picked and grown on two new LB plates at 37 °C in the dark. The growth condition of the bacteria on each plate was observed after 12-16 h. We found that for the strain containing pDawn(cI-LVA)-RBS070-LKD-pSEVA331, a significant proportion of the recombinant colonies did not grow very well in the dark, indicating a leak expression of the lysis genes. Nevertheless, for the strain containing pDawn(cI)-RBS070-LKD-pSEVA331, all the colonies grow normally in the dark. Therefore, the new pDawn in which the LVA tag of cI is deleted reduces the fitness cost of the blue light responsive lysis system, while the system with the original pDawn confers a significant growth disadvantage.
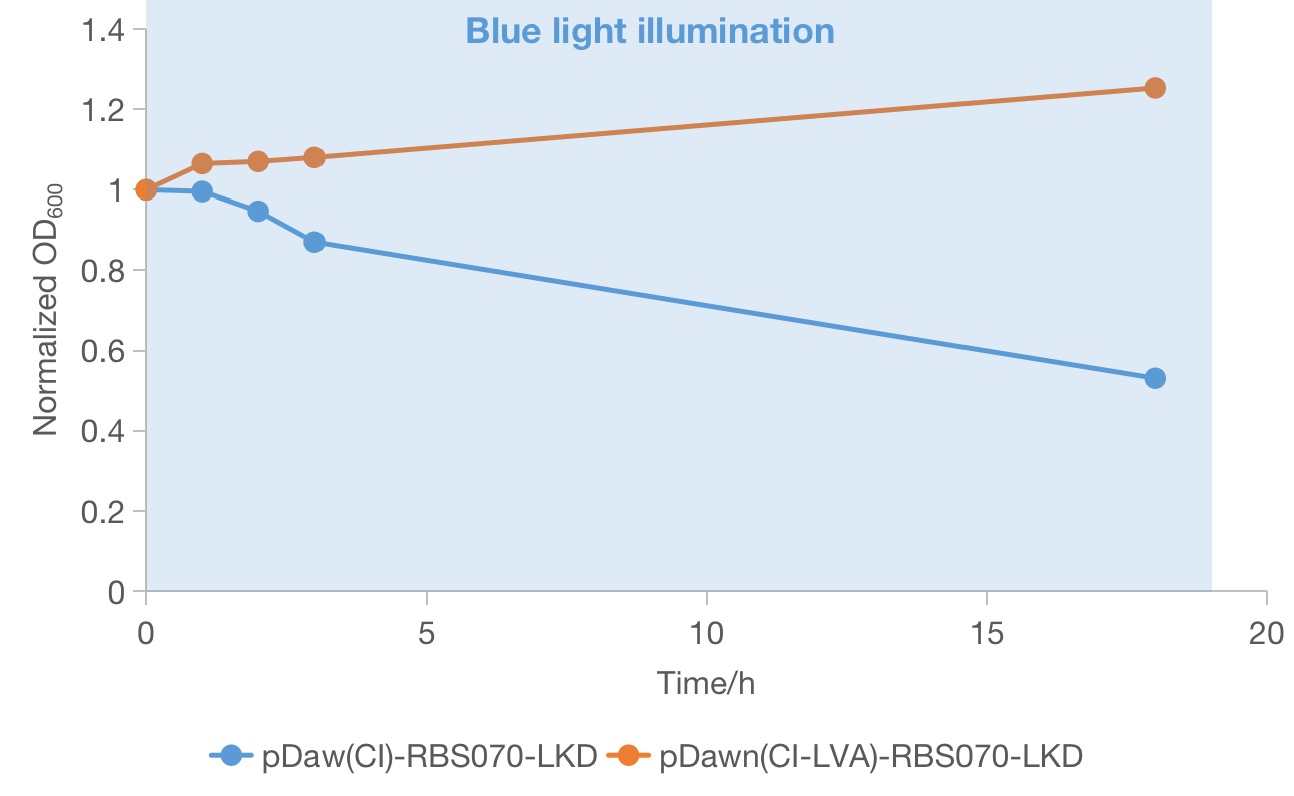
From the above two groups of successful recombinant candidates (red squares in Figure 1), one colony was picked and grow in LB medium in the dark until ~OD600 0.6 was reached to track the growth condition under blue light. Then the OD600 values of the two selected strains under blue light was measured to compare the effectiveness of the two systems in rapidly growing cells. We found that both of the strain continued to grow during the first few hours of blue light illumination. However, only the strain containing the plasmid of pDawn(cI)-RBS070-LKD-pSEVA331 showed a significant decrease in OD600 value, indicating maintenance of lethality. And the OD600 value of the strain containing the plasmid of pDawn(cI)-RBS070-LKD-pSEVA331 increases a little bit, implying that loss-of-function mutations could occurred in these genes or in the host genome and that the favorable mutant overtakes the parental population. Therefore, the new pDawn in which the LVA tag of cI is deleted increases the evolutionary stability of the blue light sensitive kill switch by negating the deleterious evolutionary pressure of leaky toxin expression.