Difference between revisions of "Part:BBa K4166004"
Djcamenares (Talk | contribs) |
Djcamenares (Talk | contribs) (→Plasmid Copy Number and Expression - Alma 2022) |
||
| Line 7: | Line 7: | ||
== Plasmid Copy Number and Expression - Alma 2022 == | == Plasmid Copy Number and Expression - Alma 2022 == | ||
| − | + | This year, we planned two improvements to the commonly used part, J04450. To better control the sometimes leaky expression, or to maximize expression, we created mutants of the origin sequence present on all pSB1C3 backbones. We introduced mutations +1, +8, -11, -13 from Camps et al, 2010, and the A715G reversion mutant from Lin-Chao et al, 1992 (turning this back into pBR322, instead of pUC19). | |
| − | + | ||
| − | We | + | |
| + | We collected plasmid copy number information using a qPCR protocol, and different approaches to gel quantitation (such as those found in Lin-Chao et al 1992 (and the variation present on the Barrick lab protocols page), Pushnova et al 2000. Here are some representative results: | ||
[[File:T--Alma--gel_004.png|350px|right]] | [[File:T--Alma--gel_004.png|350px|right]] | ||
| Line 17: | Line 16: | ||
From the intensity of these bands we estimate the copy number of this origin at ~307 copies/cell. | From the intensity of these bands we estimate the copy number of this origin at ~307 copies/cell. | ||
We found that, although this mutation increases plasmid copy number, it actually decreases expression. It could be due to collisions between DNA polymerase (attempting to replicate the plasmid) and RNA polymerase (attempting to transcribe the RFP gene). | We found that, although this mutation increases plasmid copy number, it actually decreases expression. It could be due to collisions between DNA polymerase (attempting to replicate the plasmid) and RNA polymerase (attempting to transcribe the RFP gene). | ||
| + | |||
| + | Based on several methods, we estimated the following ratio of plasmid to genomic DNA in a cell: | ||
| + | |||
| + | J04450: 76 | ||
| + | K4166002: 107 (Lin-Chao A715G) | ||
| + | K4166004: 307 (Camp +1) | ||
| + | K4166005: 381 (Camp +8) | ||
| + | K4166006: 133 (Camp +8 and -11) | ||
| + | K4166007: 153 (Camp -11) | ||
| + | K4166008: 194 (Camp +13) | ||
| + | |||
| + | In exponential cells, there is typically more than one copy of the genome present per cell, since the cells are actively replicating. Therefore, we would except to double or triple the above numbers to get a true plasmid copy number per cell. (This is based on the data at found from the BIONUMB3R5 database: https://bionumbers.hms.harvard.edu/bionumber.aspx?id=106438&ver=2&trm=Escherichia+Coli+Genome&org=) | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 00:57, 14 October 2022
Camp +1 mutation.
Increased plasmid copy number. Increased P2 extension, stabilizes secondary structure in promoter.
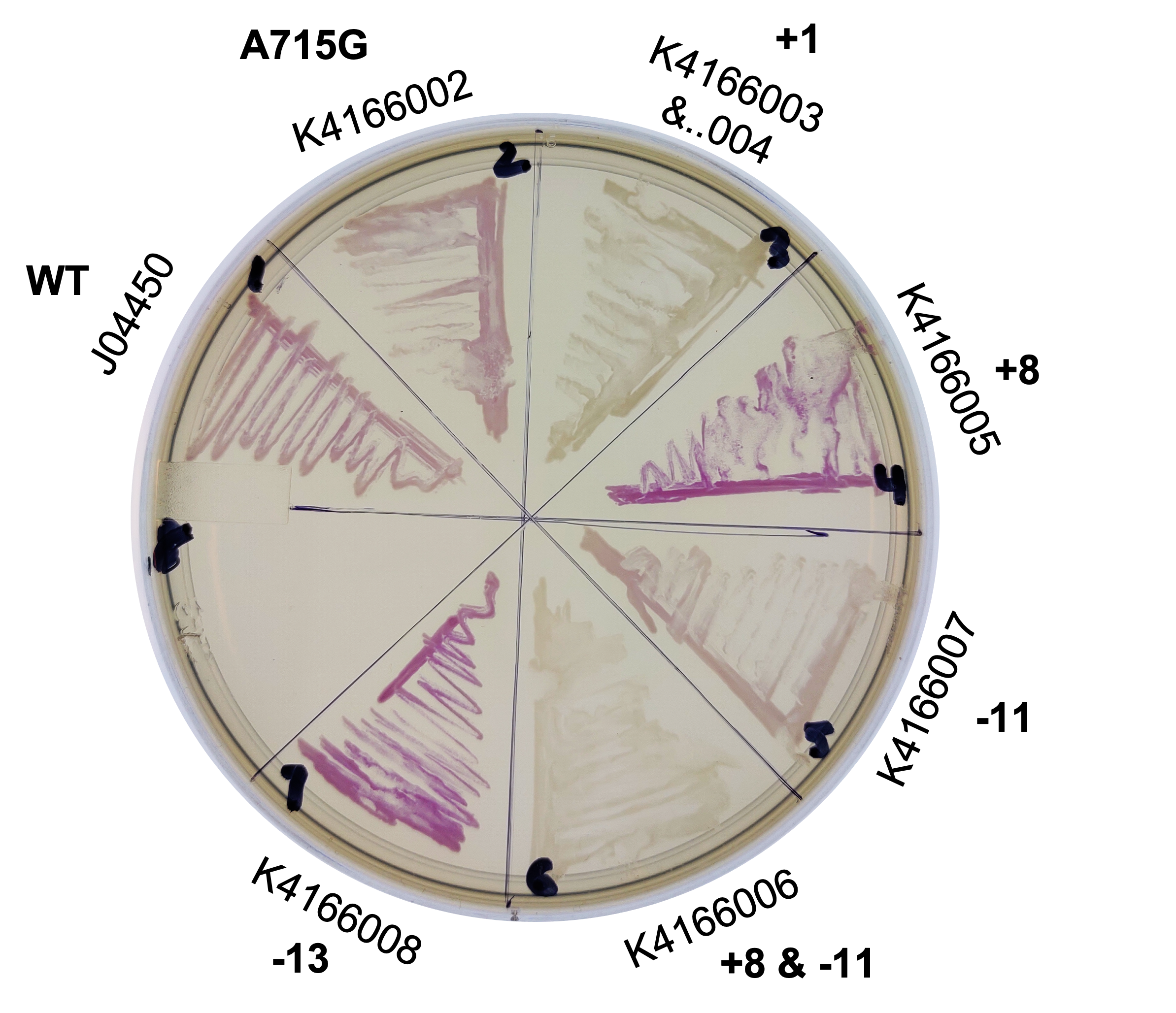
Plasmid Copy Number and Expression - Alma 2022
This year, we planned two improvements to the commonly used part, J04450. To better control the sometimes leaky expression, or to maximize expression, we created mutants of the origin sequence present on all pSB1C3 backbones. We introduced mutations +1, +8, -11, -13 from Camps et al, 2010, and the A715G reversion mutant from Lin-Chao et al, 1992 (turning this back into pBR322, instead of pUC19).
We collected plasmid copy number information using a qPCR protocol, and different approaches to gel quantitation (such as those found in Lin-Chao et al 1992 (and the variation present on the Barrick lab protocols page), Pushnova et al 2000. Here are some representative results:
From the intensity of these bands we estimate the copy number of this origin at ~307 copies/cell. We found that, although this mutation increases plasmid copy number, it actually decreases expression. It could be due to collisions between DNA polymerase (attempting to replicate the plasmid) and RNA polymerase (attempting to transcribe the RFP gene).
Based on several methods, we estimated the following ratio of plasmid to genomic DNA in a cell:
J04450: 76 K4166002: 107 (Lin-Chao A715G) K4166004: 307 (Camp +1) K4166005: 381 (Camp +8) K4166006: 133 (Camp +8 and -11) K4166007: 153 (Camp -11) K4166008: 194 (Camp +13)
In exponential cells, there is typically more than one copy of the genome present per cell, since the cells are actively replicating. Therefore, we would except to double or triple the above numbers to get a true plasmid copy number per cell. (This is based on the data at found from the BIONUMB3R5 database: https://bionumbers.hms.harvard.edu/bionumber.aspx?id=106438&ver=2&trm=Escherichia+Coli+Genome&org=)
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Plasmid lacks a prefix.
Plasmid lacks a suffix. - 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix. - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix. - 23INCOMPATIBLE WITH RFC[23]Plasmid lacks a prefix.
Plasmid lacks a suffix. - 25INCOMPATIBLE WITH RFC[25]Plasmid lacks a prefix.
Plasmid lacks a suffix. - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.