Difference between revisions of "Part:BBa K4090010"
| Line 6: | Line 6: | ||
<!-- Add more about the biology of this part here --> | <!-- Add more about the biology of this part here --> | ||
| − | + | [[File:T--SDSZ China--result inpsilicatein.png|700px|thumb|center|Fig.1]] | |
| + | [[File:T--SDSZ China--result 1-3parts.png|700px|thumb|center|Fig.2]] | ||
==Data== | ==Data== | ||
| − | [[File:T--SDSZ_China--result_1-2parts.png|700px|thumb|center|Fig. | + | [[File:T--SDSZ_China--result_1-2parts.png|700px|thumb|center|Fig.3]] |
According to Fig.1, all the fragments designed have been successfully made, including S1&S2, which are Silicatein combined with INP, and P1&P2, which are S010_pET28b-csgA-linker-mfp5-7His(pET). | According to Fig.1, all the fragments designed have been successfully made, including S1&S2, which are Silicatein combined with INP, and P1&P2, which are S010_pET28b-csgA-linker-mfp5-7His(pET). | ||
| − | [[File:T--SDSZ China--result 1-4parts.png|700px|thumb|center|Fig. | + | [[File:T--SDSZ China--result 1-4parts.png|700px|thumb|center|Fig.4]] |
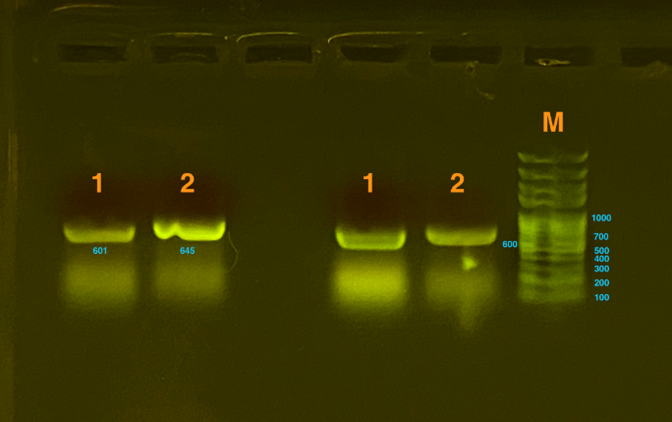
Fig.2 represents the results of gel electrophoresis to test the fragment after Gibson Assembly. The length is about 600bp, which, together with the results from sequencing, indicates a positive outcome. | Fig.2 represents the results of gel electrophoresis to test the fragment after Gibson Assembly. The length is about 600bp, which, together with the results from sequencing, indicates a positive outcome. | ||
| − | [[File:T--SDSZ China--result 1-6parts.png|700px|thumb|center|Fig. | + | [[File:T--SDSZ China--result 1-6parts.png|700px|thumb|center|Fig.5]] |
Fig.3 represents qualitative OD values, showing that there is sufficient amount of silicatein produced, since: | Fig.3 represents qualitative OD values, showing that there is sufficient amount of silicatein produced, since: | ||
•Both NC Control are darker (NC1 remain as yellow which indicates that it needs MORE NaOH to turn blue, it contains more acids). | •Both NC Control are darker (NC1 remain as yellow which indicates that it needs MORE NaOH to turn blue, it contains more acids). | ||
•Both silicatein mixture (with / without homogenization) have higher OD810nm, meaning that there are more bacteria inside the test tube (the solution is thicker / not as clear as NCs). | •Both silicatein mixture (with / without homogenization) have higher OD810nm, meaning that there are more bacteria inside the test tube (the solution is thicker / not as clear as NCs). | ||
| − | [[File:T--SDSZ China--result 1-7parts.jpeg|700px|thumb|center|Fig. | + | [[File:T--SDSZ China--result 1-7parts.jpeg|700px|thumb|center|Fig.6]] |
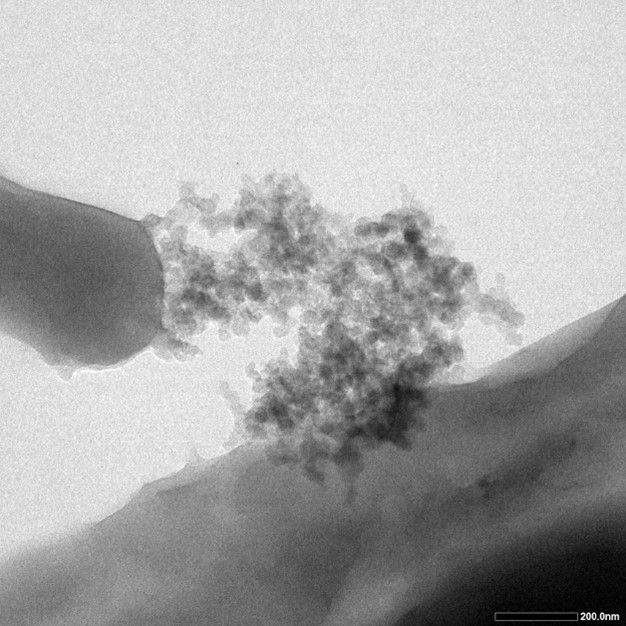
Fig.4 shows the results from TEM testing. Dark proportions represent silicon dioxide produced on the surface of cells. | Fig.4 shows the results from TEM testing. Dark proportions represent silicon dioxide produced on the surface of cells. | ||
| − | |||
| − | |||
<!-- --> | <!-- --> | ||
Revision as of 12:32, 17 October 2021
INP-Silicatein
A kind of fusion protein that helps to form silicon dioxide outside the membrane.
Data
According to Fig.1, all the fragments designed have been successfully made, including S1&S2, which are Silicatein combined with INP, and P1&P2, which are S010_pET28b-csgA-linker-mfp5-7His(pET).
Fig.2 represents the results of gel electrophoresis to test the fragment after Gibson Assembly. The length is about 600bp, which, together with the results from sequencing, indicates a positive outcome.
Fig.3 represents qualitative OD values, showing that there is sufficient amount of silicatein produced, since: •Both NC Control are darker (NC1 remain as yellow which indicates that it needs MORE NaOH to turn blue, it contains more acids). •Both silicatein mixture (with / without homogenization) have higher OD810nm, meaning that there are more bacteria inside the test tube (the solution is thicker / not as clear as NCs).
Fig.4 shows the results from TEM testing. Dark proportions represent silicon dioxide produced on the surface of cells.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1392
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 321
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 63
Illegal NgoMIV site found at 396
Illegal AgeI site found at 498 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 229
References
[1] Pamirsky, Igor E., and Kirill S. Golokhvast. "Silaffins of diatoms: from applied biotechnology to biomedicine." Marine drugs 11.9 (2013): 3155-3167.
[2] Park, Jeong Chan, et al. "R5 Peptide-based Biosilicification Using Methyltrimethoxysilane." Biotechnology & Bioprocess Engineering 23.1 (2018).
[3] Müller, Werner EG, et al. "Bioencapsulation of living bacteria (Escherichia coli) with poly (silicate) after transformation with silicatein-α gene." Biomaterials 29.7 (2008): 771-779.
[4] ZHU T, PAULO C, MERROUN, M L, et al. Potential application of biomineralization by Synechococcus PCC8806 for concrete restoration[J]. Sedimentology, 2013,61(1):1-21.
[5] Lechner, Carolin C., and Christian FW Becker. "A sequence‐function analysis of the silica precipitating silaffin R5 peptide." Journal of Peptide Science 20.2 (2014): 152-158.