Difference between revisions of "Part:BBa K3887010"
| Line 3: | Line 3: | ||
<partinfo>BBa_K3887010 short</partinfo> | <partinfo>BBa_K3887010 short</partinfo> | ||
| − | + | This is a plasmid designed to knock out the TnaA gene on E. coli genome. Its core part is an RNA scaffold under the control of the J23119 promoter, sgRNA and the homology arm sequence at both ends of the TnaA gene (Figure 1). | |
| + | [[File:T--BJU_China--Fig.1 target1.png|600px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Figure.1 Gene circuit of pTarget-ΔTnaA</center></b></p> | ||
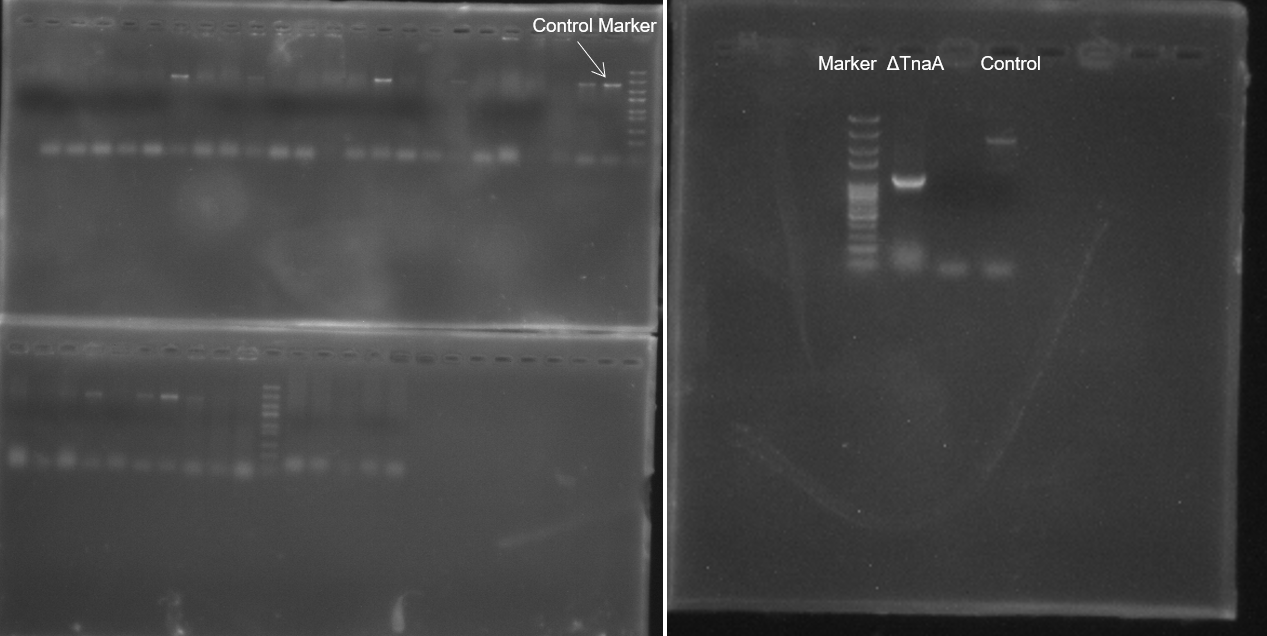
| + | In the original design of this plasmid, it was resistant to ampicillin antibiotics. We did many experiments and found that all the selected colonies failed to knock out the TnaA gene (Figure 2, left). After analysis, we suspected that it may due to the special antibacterial principle of ampicillin, the effective ampicillin in the plate medium reduced over time, which will result in the ineffective screening of the plate and obtain multiple false positive colonies. | ||
| + | For this reason, we replaced the ampicillin antibiotic gene in the plasmid with the chloramphenicol gene, and finally successfully knocked out the TnaA gene. Through the results (Figure 2, right),we can obviously find the PCR product length was short when compared to control group using the same primers. | ||
| + | [[File:T--BJU_China--Fig. 2 target2.png|600px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Figure.2 Colony PCR results of pTarget-ΔTnaA with different resistance</center></b></p> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 09:53, 20 October 2021
pTarget-ΔTnaA
This is a plasmid designed to knock out the TnaA gene on E. coli genome. Its core part is an RNA scaffold under the control of the J23119 promoter, sgRNA and the homology arm sequence at both ends of the TnaA gene (Figure 1).
In the original design of this plasmid, it was resistant to ampicillin antibiotics. We did many experiments and found that all the selected colonies failed to knock out the TnaA gene (Figure 2, left). After analysis, we suspected that it may due to the special antibacterial principle of ampicillin, the effective ampicillin in the plate medium reduced over time, which will result in the ineffective screening of the plate and obtain multiple false positive colonies. For this reason, we replaced the ampicillin antibiotic gene in the plasmid with the chloramphenicol gene, and finally successfully knocked out the TnaA gene. Through the results (Figure 2, right),we can obviously find the PCR product length was short when compared to control group using the same primers.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1335
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 150
Illegal BamHI site found at 1347
Illegal XhoI site found at 126 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 1410