Difference between revisions of "Part:BBa K3796220"
Yang Feiyue (Talk | contribs) |
|||
| Line 8: | Line 8: | ||
Used to measure the strength of P0864, which is a blank control. | Used to measure the strength of P0864, which is a blank control. | ||
P0864 is an endogenous strong promoter in the genome of <i>Corynebacterium glutamicum</i> ATCC13032. This promoter is linked upstream of the gene and can be expressed constitutively in <i>Corynebacterium glutamicum</i> with high expression intensity by transformation into it. | P0864 is an endogenous strong promoter in the genome of <i>Corynebacterium glutamicum</i> ATCC13032. This promoter is linked upstream of the gene and can be expressed constitutively in <i>Corynebacterium glutamicum</i> with high expression intensity by transformation into it. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
===Characterization=== | ===Characterization=== | ||
| − | + | <p>This part is the genetic circuit we used as blank control to test the strength of P0864 in <i>C. glutamicum</i> and <i>E. coli</i>.</p> | |
| − | <p> | + | <p>We want to solve this by inserting a reporter gene behind P0864 to visually show its strength as a promoter. For convenience, we choose the fluorescent protein mCherry, whose excitation light and emission light both fall into the range of visible light. Also, we choose to use the shuttle vector pXMJ19 that is applicable to both <i>E. coli</i> and <i>C. glutamicum</i>. The blank control we used is BBa_K3796220. Particularly, you may notice that there is a tac promoter upstream of the MCS on pXMJ19, so a proper blank control to exclude the influence of Ptac leaky expression is needed. Ideally, the fluorescence intensity/OD<sub>600</sub> should be higher in experiment group than in the control group. </p> |
| − | [[image:T--CAU China-- | + | [[image:T--CAU China--Part01 pmc.svg|600px|thumb|center|]] |
| − | <p style="text-align: center;"><b>Fig.1 | + | <p style="text-align: center;"><b>Fig.1 Experimental group genetic circuit(BBa_K3796219)</b></p> |
<br> | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
[[image:T--CAU China--Part02 mc.svg|600px|thumb|center|]] | [[image:T--CAU China--Part02 mc.svg|600px|thumb|center|]] | ||
| − | <p style="text-align: center;"><b>Fig. | + | <p style="text-align: center;"><b>Fig.2 Control group genetic circuit(BBa_K3796220)</b></p> |
<br> | <br> | ||
| + | <p> | ||
| + | We built the genetic circuits using Clonexpress® Multi One Step Cloning kit from Vazyme to connect RBS sequence (BBa_K3796001, synthetic) and mCherry gene (BBa_J06504) downstream to P0864. We successfully constructed the above sequences and verified them by colony PCR and sequencing.</p> | ||
<p>The transformed strains were cultured on LBG chloramphenicol resistant solid medium for 24 hours. In <i>E.coli</i>, two gene circuits showed different fluorescence. It has different performance under the irradiation of different light sources. The fluorescence of circuit containing P0864 is significantly brighter than that of another circuit. Under the irradiation of natural light, the colonies showed purplish red.</p> | <p>The transformed strains were cultured on LBG chloramphenicol resistant solid medium for 24 hours. In <i>E.coli</i>, two gene circuits showed different fluorescence. It has different performance under the irradiation of different light sources. The fluorescence of circuit containing P0864 is significantly brighter than that of another circuit. Under the irradiation of natural light, the colonies showed purplish red.</p> | ||
[[image:T--CAU China--Part03 expr&ck.png|400px|thumb|center|]] | [[image:T--CAU China--Part03 expr&ck.png|400px|thumb|center|]] | ||
| − | <p style="text-align: center;"><b>Fig. | + | <p style="text-align: center;"><b>Fig.3 The difference of fluorescent protein expressed by positive <i>Escherichia coli</i> (including experimental group and control group). a. Image under excitation light.(Left: Experimental group, right: control group) b. Images in natural light.(Left: Experimental group, right: control group)</b></p> |
| − | <br> | + | <br><br> |
| − | < | + | |
[[image:T--CAU China--Parts tube.png|400px|thumb|center|]] | [[image:T--CAU China--Parts tube.png|400px|thumb|center|]] | ||
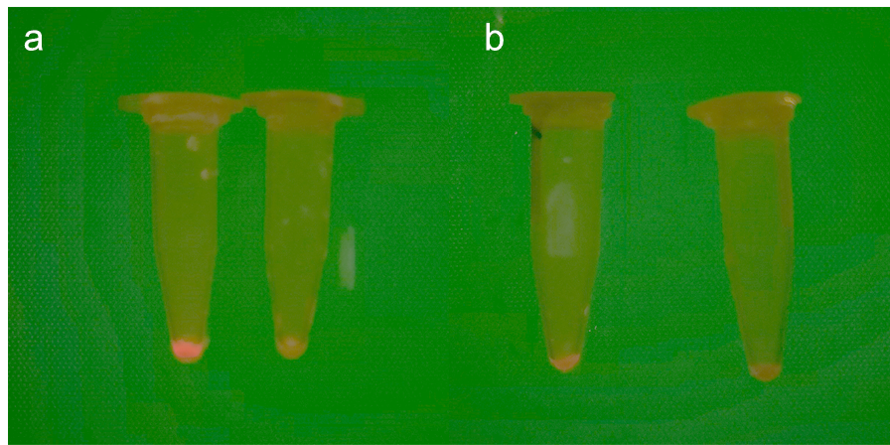
| − | <p style="text-align: center;"><b>Fig. | + | <p style="text-align: center;"><b>Fig.4 Difference of fluorescent proteins expressed by positive <i>Escherichia coli</i> and <i>Corynebacterium glutamicum</i> a. Images of <i>Corynebacterium glutamicum</i> after centrifugation, including experimental group (left) and control group (right). b. Images of <i>E. coli</i> after centrifugation, including experimental group (left) and control group (right).</b></p> |
| + | |||
| + | <p> | ||
| + | We designed a quantitative test to better characterize this promoter in different chassis. After the bacteria had been cultured at 37 ℃ for 26 hours, the fluorescence was measured with a fluorescence spectrophotometer and OD<sub>600</sub> was measured with a visible spectrophotometer. We use the ratio of fluorescence intensity/OD<sub>600</sub> to reflect the relative fluorescence intensity under the unit absorbance value. | ||
| + | </p> | ||
<br> | <br> | ||
[[image:T--CAU China--Parts data.png|400px|thumb|center|]] | [[image:T--CAU China--Parts data.png|400px|thumb|center|]] | ||
| − | <p style="text-align: center;"><b>Fig.6 After <i>E.coli</i> and <i>C.glutamicum</i> were cultured at | + | <p style="text-align: center;"><b>Fig.6 After <i>E.coli</i> and <i>C.glutamicum</i> were cultured at 37 ℃ for 26 hours, the ratio of measured fluorescence data to OD600 data.</b></p> |
<br> | <br> | ||
<p>T-test was performed on the three groups of data.</p> | <p>T-test was performed on the three groups of data.</p> | ||
| − | <p><b>Table.1 T-test results of two species. ***: p-value<0.001, **: p-value<0.01</b></p> | + | <p><b><center>Table.1 T-test results of two species. ***: p-value<0.001, **: p-value<0.01</center></b></p> |
[[image:T--CAU China--Parts ttest.png|400px|thumb|center|]] | [[image:T--CAU China--Parts ttest.png|400px|thumb|center|]] | ||
| − | <p>The mean value of experimental group was significantly different from that of control group. | + | <p>The mean value of experimental group was significantly different from that of control group. We learned that P0864 is a constitutive promoter that works both in <i>E. coli</i> and <i>C. glutamicum</i>. Its strength tends to be stronger in <i>C. glutamicum</i>. If used in <i>E. coli </i>, codon-optimization may be carried out.To sum up, by using BBa_K3796219 and BBa_K3796220 as blank control, we knew that P0864 can be used as a constitutive promoter in future projects.</p> |
| − | + | ||
Revision as of 03:22, 21 October 2021
P0864 strength measurement blank
Used to measure the strength of P0864, which is a blank control.
P0864 is an endogenous strong promoter in the genome of Corynebacterium glutamicum ATCC13032. This promoter is linked upstream of the gene and can be expressed constitutively in Corynebacterium glutamicum with high expression intensity by transformation into it.
Characterization
This part is the genetic circuit we used as blank control to test the strength of P0864 in C. glutamicum and E. coli.
We want to solve this by inserting a reporter gene behind P0864 to visually show its strength as a promoter. For convenience, we choose the fluorescent protein mCherry, whose excitation light and emission light both fall into the range of visible light. Also, we choose to use the shuttle vector pXMJ19 that is applicable to both E. coli and C. glutamicum. The blank control we used is BBa_K3796220. Particularly, you may notice that there is a tac promoter upstream of the MCS on pXMJ19, so a proper blank control to exclude the influence of Ptac leaky expression is needed. Ideally, the fluorescence intensity/OD600 should be higher in experiment group than in the control group.
Fig.1 Experimental group genetic circuit(BBa_K3796219)
Fig.2 Control group genetic circuit(BBa_K3796220)
We built the genetic circuits using Clonexpress® Multi One Step Cloning kit from Vazyme to connect RBS sequence (BBa_K3796001, synthetic) and mCherry gene (BBa_J06504) downstream to P0864. We successfully constructed the above sequences and verified them by colony PCR and sequencing.
The transformed strains were cultured on LBG chloramphenicol resistant solid medium for 24 hours. In E.coli, two gene circuits showed different fluorescence. It has different performance under the irradiation of different light sources. The fluorescence of circuit containing P0864 is significantly brighter than that of another circuit. Under the irradiation of natural light, the colonies showed purplish red.
Fig.3 The difference of fluorescent protein expressed by positive Escherichia coli (including experimental group and control group). a. Image under excitation light.(Left: Experimental group, right: control group) b. Images in natural light.(Left: Experimental group, right: control group)
Fig.4 Difference of fluorescent proteins expressed by positive Escherichia coli and Corynebacterium glutamicum a. Images of Corynebacterium glutamicum after centrifugation, including experimental group (left) and control group (right). b. Images of E. coli after centrifugation, including experimental group (left) and control group (right).
We designed a quantitative test to better characterize this promoter in different chassis. After the bacteria had been cultured at 37 ℃ for 26 hours, the fluorescence was measured with a fluorescence spectrophotometer and OD600 was measured with a visible spectrophotometer. We use the ratio of fluorescence intensity/OD600 to reflect the relative fluorescence intensity under the unit absorbance value.
Fig.6 After E.coli and C.glutamicum were cultured at 37 ℃ for 26 hours, the ratio of measured fluorescence data to OD600 data.
T-test was performed on the three groups of data.
The mean value of experimental group was significantly different from that of control group. We learned that P0864 is a constitutive promoter that works both in E. coli and C. glutamicum. Its strength tends to be stronger in C. glutamicum. If used in E. coli , codon-optimization may be carried out.To sum up, by using BBa_K3796219 and BBa_K3796220 as blank control, we knew that P0864 can be used as a constitutive promoter in future projects.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]