Difference between revisions of "Part:BBa K3796206"
Yang Feiyue (Talk | contribs) |
|||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3796206 short</partinfo> | <partinfo>BBa_K3796206 short</partinfo> | ||
| − | This part is composed of an inducible promoter PgsiB, a rbs sequence in Corynebacterium glutamicum (BBa_K2963006), the fluorescent protein gene bjGFP, and the rrnB T1 and T2 terminators (BBa_K3568009). | + | This part is composed of an inducible promoter PgsiB, a rbs sequence in Corynebacterium glutamicum (BBa_K2963006), the fluorescent protein gene <i>bjGFP</i>, and the rrnB T1 and T2 terminators (BBa_K3568009). PgsiB is a sigma factor B dependent promoter from <i>Bacillus subtilis</i> and can be activated under various mild stress, including salt, alkaline, heat shock, ethanol and glucose limitation. Under these stresses, the <i>gfp</i> will have higher expression, leading to stronger fluorescence intensity. |
| − | <!-- Add more about the biology of this part here | + | <!-- Add more about the biology of this part here--> |
| − | === | + | ===Characterization=== |
| + | <p>CAU_China 2021 used this part in their composite kill switch to make sure the engineered bacteria would die when the salinity and alkalinity of the soil decrease to an expected level. Since the promoter PgsiB can be activated under mild stress, we use it to respond to salt stress in <i>Corynebacterium glutamicum</i> in our project.</p> | ||
| + | <p>Hence, we aim to verify that the promoter PgsiB, originally from <i> Bacillus subtilis</i>, can respond to salt stress in <i>Corynebacterium glutamicum</i> and we also want to document its response towards different salt concentration.</p> | ||
| + | <p>We use the gene circuit below(BBa_K3796206) to test the function of PgsiB by the fluorescence of GFP.</p> | ||
| + | <br> | ||
| + | |||
| + | [[File:T--CAU_China--PgsiB_Circuit.svg|600px|thumb|center|]] | ||
| + | <p style="text-align: center;"><b>Fig.1 Genetic circuit for PgsiB verification</b></p> | ||
| + | |||
| + | <br> | ||
| + | <p>We build the gene circuit by ClonExpress II one-step cloning kit (Vazyme Biotech, China). Particularly, we destroyed the tac promoter on the plasmid pXMJ19 to get rid of its influence.</p> | ||
| + | <p>After sequencing and amplifying, that vector as well as unmodified pXMJ19 were transferred into <i>Corynebacterium glutamicum</i> separately and cultivated on plates. After cultivating them in the LB liquid culture medium at 100 rpm, 30℃ for 12h, the OD<SUB>600</SUB> of the two bacteria were adjusted to nearly the same(nearly 2.0) and inoculated in the salt gradient LB liquid medium. After a cultivation of 24h, the bacterial solution was collected and washed with PBS. Then we measured its fluorescence intensity by HITACHI F-7000, a fluorescence spectrophotometer, according to the excitation light of 488nm and emission light of 507nm and OD<SUB>600</SUB>. Data are shown below:</p> | ||
| + | <br> | ||
| + | |||
| + | [[File:T--CAU_China--PgsiB_Result.png|600px|thumb|center|]] | ||
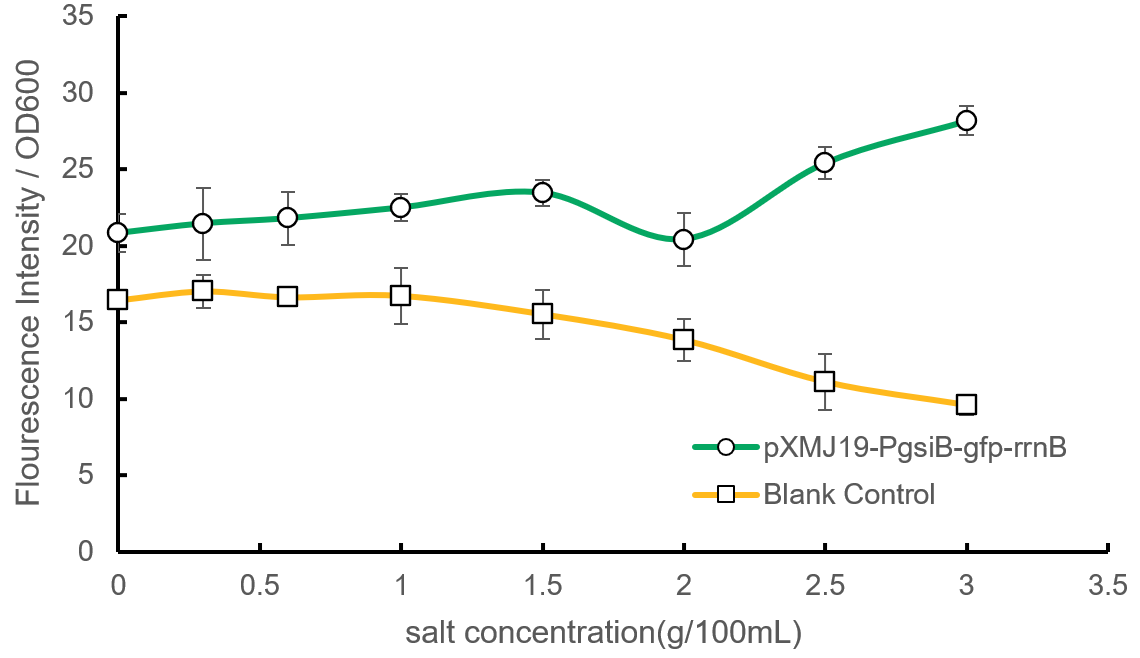
| + | <p style="text-align: center;"><b>Fig.2 Fluorescence intensity results in the verification of PgsiB (the excitation light of 488nm and emission light of 507nm)</b></p> | ||
| + | |||
| + | <p>We use the control group(bacteria with empty vector) to exclude the influence of the fluorescence bacteria originally have. Through two-way ANOVA, we know there is significant difference between the control group and the experiment group, proving our results valid. Data shows that the difference of relative fluorescence intensity(fluorescence intensity/OD600) between the two becomes bigger as the salt concentration increase, indicating that there is higher expression of <i>gfp</i> under the control of PgsiB when the salt concentration is high.</p> | ||
| + | <p>We finally come to the conclusion that <strong>PgsiB can respond to salinity changes in <i>C.glutamicum</i>, and high salt concentration can improve the expression of the downstream genes of PgsiB.</strong> Three parallel experiments were done later, which supported our conclusion. However, we haven’t observed a typical turning point in our curves, and we want to do more experiments about that in near future.</p> | ||
| + | |||
| + | <br><br> | ||
<!-- --> | <!-- --> | ||
Revision as of 01:57, 21 October 2021
PgsiB-rbs-gfp-terminator
This part is composed of an inducible promoter PgsiB, a rbs sequence in Corynebacterium glutamicum (BBa_K2963006), the fluorescent protein gene bjGFP, and the rrnB T1 and T2 terminators (BBa_K3568009). PgsiB is a sigma factor B dependent promoter from Bacillus subtilis and can be activated under various mild stress, including salt, alkaline, heat shock, ethanol and glucose limitation. Under these stresses, the gfp will have higher expression, leading to stronger fluorescence intensity.
Characterization
CAU_China 2021 used this part in their composite kill switch to make sure the engineered bacteria would die when the salinity and alkalinity of the soil decrease to an expected level. Since the promoter PgsiB can be activated under mild stress, we use it to respond to salt stress in Corynebacterium glutamicum in our project.
Hence, we aim to verify that the promoter PgsiB, originally from Bacillus subtilis, can respond to salt stress in Corynebacterium glutamicum and we also want to document its response towards different salt concentration.
We use the gene circuit below(BBa_K3796206) to test the function of PgsiB by the fluorescence of GFP.
Fig.1 Genetic circuit for PgsiB verification
We build the gene circuit by ClonExpress II one-step cloning kit (Vazyme Biotech, China). Particularly, we destroyed the tac promoter on the plasmid pXMJ19 to get rid of its influence.
After sequencing and amplifying, that vector as well as unmodified pXMJ19 were transferred into Corynebacterium glutamicum separately and cultivated on plates. After cultivating them in the LB liquid culture medium at 100 rpm, 30℃ for 12h, the OD600 of the two bacteria were adjusted to nearly the same(nearly 2.0) and inoculated in the salt gradient LB liquid medium. After a cultivation of 24h, the bacterial solution was collected and washed with PBS. Then we measured its fluorescence intensity by HITACHI F-7000, a fluorescence spectrophotometer, according to the excitation light of 488nm and emission light of 507nm and OD600. Data are shown below:
Fig.2 Fluorescence intensity results in the verification of PgsiB (the excitation light of 488nm and emission light of 507nm)
We use the control group(bacteria with empty vector) to exclude the influence of the fluorescence bacteria originally have. Through two-way ANOVA, we know there is significant difference between the control group and the experiment group, proving our results valid. Data shows that the difference of relative fluorescence intensity(fluorescence intensity/OD600) between the two becomes bigger as the salt concentration increase, indicating that there is higher expression of gfp under the control of PgsiB when the salt concentration is high.
We finally come to the conclusion that PgsiB can respond to salinity changes in C.glutamicum, and high salt concentration can improve the expression of the downstream genes of PgsiB. Three parallel experiments were done later, which supported our conclusion. However, we haven’t observed a typical turning point in our curves, and we want to do more experiments about that in near future.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]