Difference between revisions of "Part:BBa K3595016"
Liyingying (Talk | contribs) |
|||
| Line 5: | Line 5: | ||
it | it | ||
| − | <!-- Add more about the biology of this part here | + | <!-- Add more about the biology of this part here--> |
| − | + | . | |
| + | =Usage and Biology= | ||
| + | This part is a coding sequence after the promoter Ptac and RBS [[Part:B0034|B0034]]. The enzyme geranyl pyrophosphate synthase can be translated under the induction of IPTG. We connected GPPS and MS with the promoter Ptac to express genes GPPS and MS on the plasmid pET28B. The constructed plasmid was transformed into the DH10b host cell to test its production of myrcene. | ||
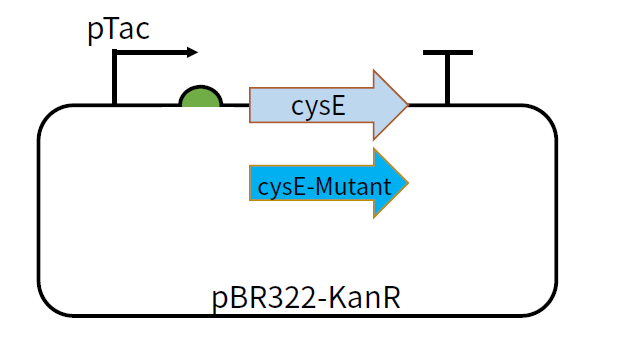
| + | [[File:T--GZ_HFI--cysE.png|600px|thumb|center|The structure of the plasmid pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-mutant ]] | ||
| + | ==Experimental Setup== | ||
| + | *Genetic information of MS was described in the page [[Part: BBa_K35950015]]. | ||
| + | *Plasmid pTYT-GPPS-MS was transferred into <i>E.coli DH10b </i> host cell,respestively. | ||
| + | *Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm. | ||
| + | *DH10b was introduced in 2 ml M9 medium with 0.5 mM IPTG and 1% glucose, in 37 degree Celsius overnight. 20% dodecane (v/v) was added to collect the myrcene produced by DH10b. | ||
| + | *After the centrifugation (12000rpm for a minute), the dodecane layer was collected to test the production of myrcene. | ||
| + | ==Results== | ||
| + | *DH10b with plasmids pMevT, pMBIS, and pTYT-GPPS-MS could produce myrcene, but in relatively low productivity compared to the previous research. | ||
| + | [[File:T--GZ_HFI--MS.png|600px|thumb|center|Detection result of the production of myrcene by the engineered bacteria. (A) GC-MS result of standard myrcene sample, blue arrow indicates the peak of myrcene. (B) GC-MS result of the culture solution of pMevT pMBIS pTYT-GPPS-MS transformed E.coli DH10b, blue arrow indicates the peak of myrcene. (C) The Calibration Line of myrcene. (D) The myrcene peak area and myrcene concentration of each group. ]] | ||
| + | |||
<!-- --> | <!-- --> | ||
Revision as of 15:22, 27 October 2020
GPPS-MS regulated by pTac
it
.
Usage and Biology
This part is a coding sequence after the promoter Ptac and RBS B0034. The enzyme geranyl pyrophosphate synthase can be translated under the induction of IPTG. We connected GPPS and MS with the promoter Ptac to express genes GPPS and MS on the plasmid pET28B. The constructed plasmid was transformed into the DH10b host cell to test its production of myrcene.
Experimental Setup
- Genetic information of MS was described in the page Part: BBa_K35950015.
- Plasmid pTYT-GPPS-MS was transferred into E.coli DH10b host cell,respestively.
- Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm.
- DH10b was introduced in 2 ml M9 medium with 0.5 mM IPTG and 1% glucose, in 37 degree Celsius overnight. 20% dodecane (v/v) was added to collect the myrcene produced by DH10b.
- After the centrifugation (12000rpm for a minute), the dodecane layer was collected to test the production of myrcene.
Results
- DH10b with plasmids pMevT, pMBIS, and pTYT-GPPS-MS could produce myrcene, but in relatively low productivity compared to the previous research.

Detection result of the production of myrcene by the engineered bacteria. (A) GC-MS result of standard myrcene sample, blue arrow indicates the peak of myrcene. (B) GC-MS result of the culture solution of pMevT pMBIS pTYT-GPPS-MS transformed E.coli DH10b, blue arrow indicates the peak of myrcene. (C) The Calibration Line of myrcene. (D) The myrcene peak area and myrcene concentration of each group.
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 2573
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 2438
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 741
Illegal AgeI site found at 1919 - 1000COMPATIBLE WITH RFC[1000]