Difference between revisions of "Part:BBa K3595002"
Liyingying (Talk | contribs) |
Liyingying (Talk | contribs) |
||
| Line 11: | Line 11: | ||
==Experimental Setup== | ==Experimental Setup== | ||
*Genetic information of argA was described on the page of [[Part:BBa_K3595001]] | *Genetic information of argA was described on the page of [[Part:BBa_K3595001]] | ||
| − | *Plasmid pBR322-KanR-pTac-argA and pBR322-KanR-pTac-argA ^ fbr was transfered into the <i>E.coli DH10b-ΔargR </i> host cell,respestively. Meanwhile,plasmid pBR322-KanR-pTac-argA was transfered into the<i>E.coli DH10b | + | *Plasmid pBR322-KanR-pTac-argA and pBR322-KanR-pTac-argA ^ fbr was transfered into the <i>E.coli DH10b-ΔargR </i> host cell,respestively. Meanwhile,plasmid pBR322-KanR-pTac-argA was transfered into the<i>E.coli DH10b </i> as a negative control, and M9 media as a blank control. |
*Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm. | *Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm. | ||
*Inoculating 15 uL of culture solution overnight into a 24-well plate containing 3 mL M9 medium for overnight growth at 37 °C and 200 rpm.. The media contained 3 ul kanamycin , 1.5 uL 1M IPTG and 5mM ammonia. | *Inoculating 15 uL of culture solution overnight into a 24-well plate containing 3 mL M9 medium for overnight growth at 37 °C and 200 rpm.. The media contained 3 ul kanamycin , 1.5 uL 1M IPTG and 5mM ammonia. | ||
Revision as of 06:14, 27 October 2020
Mutant argA-fbr
ArgA gene takes part in the first step that converts glutamate into arginine. The function of the synthetase encoded by ArgA is influenced by the concentration of arginine. The higher the concentration, the lower effect the enzyme will have.However, the arginine synthesis pathway is repressed through two regulatory pathways.One of them is arginine itself has feedback inhibition on NAGS. In order to maximize the conversion of ammonia to arginine, feedback-resistant (fbr) NAGS enzymes is required, whose argA gene is mutated into argA^fbr by substituting the sequences TAT (Tyr) at 19th position into TGT (Cys), and the GCG (Ala) at 389th position into GCT (Ala).
Usage and Biology
This part can be used as a coding sequence after the promoter pTac and RBS B0034. The argA-fbr protein can be translated under the induction of IPTG. We constructed plasmids pBR322-KanR-pTac-argA and pBR322-KanR-pTac-argA using argA and argAfbr, respectively. The constructed plasmid was transformed into E.coli DH10b-ΔargR host cell to test its ammonia degradation efficiency.
Experimental Setup
- Genetic information of argA was described on the page of Part:BBa_K3595001
- Plasmid pBR322-KanR-pTac-argA and pBR322-KanR-pTac-argA ^ fbr was transfered into the E.coli DH10b-ΔargR host cell,respestively. Meanwhile,plasmid pBR322-KanR-pTac-argA was transfered into theE.coli DH10b as a negative control, and M9 media as a blank control.
- Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm.
- Inoculating 15 uL of culture solution overnight into a 24-well plate containing 3 mL M9 medium for overnight growth at 37 °C and 200 rpm.. The media contained 3 ul kanamycin , 1.5 uL 1M IPTG and 5mM ammonia.
- Detecting ammonia concentration in culture medium
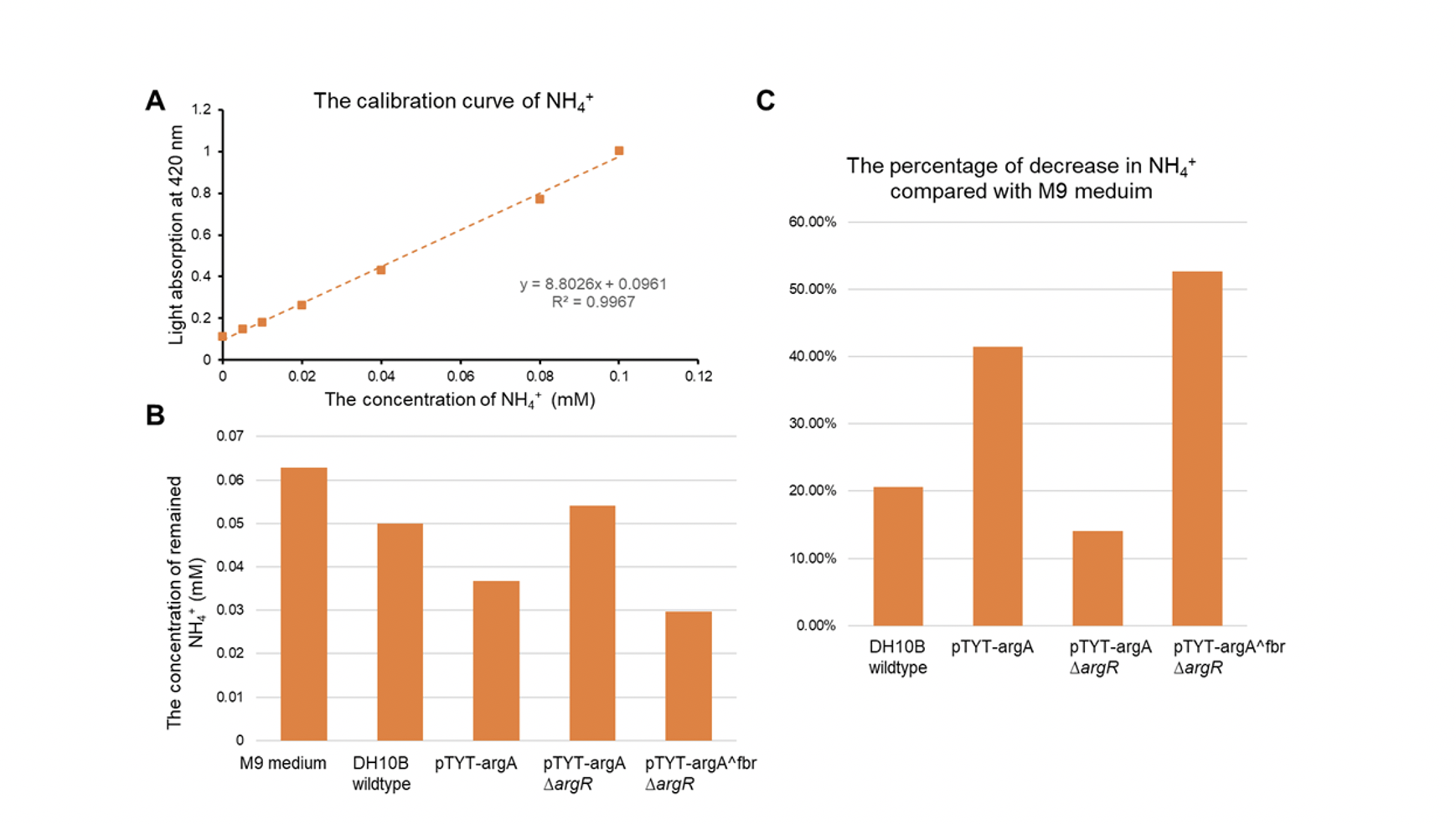
Results
- The DH10b with plasmid pTYT-argAfbr and ∆argR showed the most significant increase in conversion of ammonia, 40.47% more conversion rate compared to the wildtype
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 626