Difference between revisions of "Part:BBa K3429011"
| Line 33: | Line 33: | ||
</html> | </html> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K3429011 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3429011 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <h2>CotA Docking</h2> | ||
| + | <div class="containertext"> | ||
| + | At first we did the ligand docking of diclofenac with the laccase CotA. Preparations of the ligand and protein were done as explained above. For realization of the docking process we needed a working Rosetta xml script for our purpose. The script we worked out is shown in our <a href="https://2020.igem.org/Team:TU_Darmstadt/Model/Rosetta_Guide" target="blank">Rosetta Guide</a>. With this script, adjusted over many trials, we docked many thousand structures. As output we chose the Rosetta total score and the Rosetta interface score. We plotted these scores against the <b>root-mean-square deviation of atomic positions (RMSD)</b> from the ligand to allow a visible evaluation of the docked structures respectively poses<sup id="cite_ref-9"><a href="#cite_note-9">[9]</sup></a>. The formula for calculation of the RMSD is shown below (formula 1).<br> | ||
| + | </div> | ||
| + | |||
| + | <!---Formel----> | ||
| + | |||
| + | <div style="display: flex;justify-content: center;"> | ||
| + | <figure> | ||
| + | <a href="https://2020.igem.org/wiki/images/4/44/T--TU_Darmstadt--RMSD_Formula_2.png"><img src="https://2020.igem.org/wiki/images/4/44/T--TU_Darmstadt--RMSD_Formula_2.png" style="width:40%; margin-left:15% alt="figure" border: 1px solid;border-color: black;"> | ||
| + | </a> <figcaption id="Figure4"><b>Formula 1: RMSD formula.</b> Calculation of the RMSD value with the respective Rosetta score function<sup id="cite_ref-9"><a href="#cite_note-9">[9]</sup></a>.</figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <br> | ||
| + | |||
| + | <div class="containertext"> | ||
| + | An <b>interface score against RMSD plot</b> from Diclofenac with CotA is shown in figure 4. Based on these plots it was easier to identify low energy structures with good interactions with the enzyme. Ideally the data points of these plots should form a funnel like plot. The structures on the closing end are especially good structures. These structures are highlighted with by a red square in figure 4.<br> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <figure> | ||
| + | <a href="https://2020.igem.org/wiki/images/3/37/T--TU_Darmstadt--Interfacescore-RMSD-Plot_CotA%2BDCF_02.png" target="_blank"><img src="https://2020.igem.org/wiki/images/3/37/T--TU_Darmstadt--Interfacescore-RMSD-Plot_CotA%2BDCF_02.png" alt="figure" style="width:65%; height:65%; margin-left:15%;border: 1px solid;border-color: black;"> | ||
| + | </a> <figcaption id="Figure4"><b>Figure 4: Interface score RMSD plot of diclofenac docking with CotA.</b> RMSD values of diclofenac in nm plotted against the Rosetta interface score. Every datapoint represents a single docked structure. The structures highlighted with a red box are especially good structures.</figcaption> | ||
| + | </figure> | ||
| + | |||
| + | |||
| + | <div class="containertext"> | ||
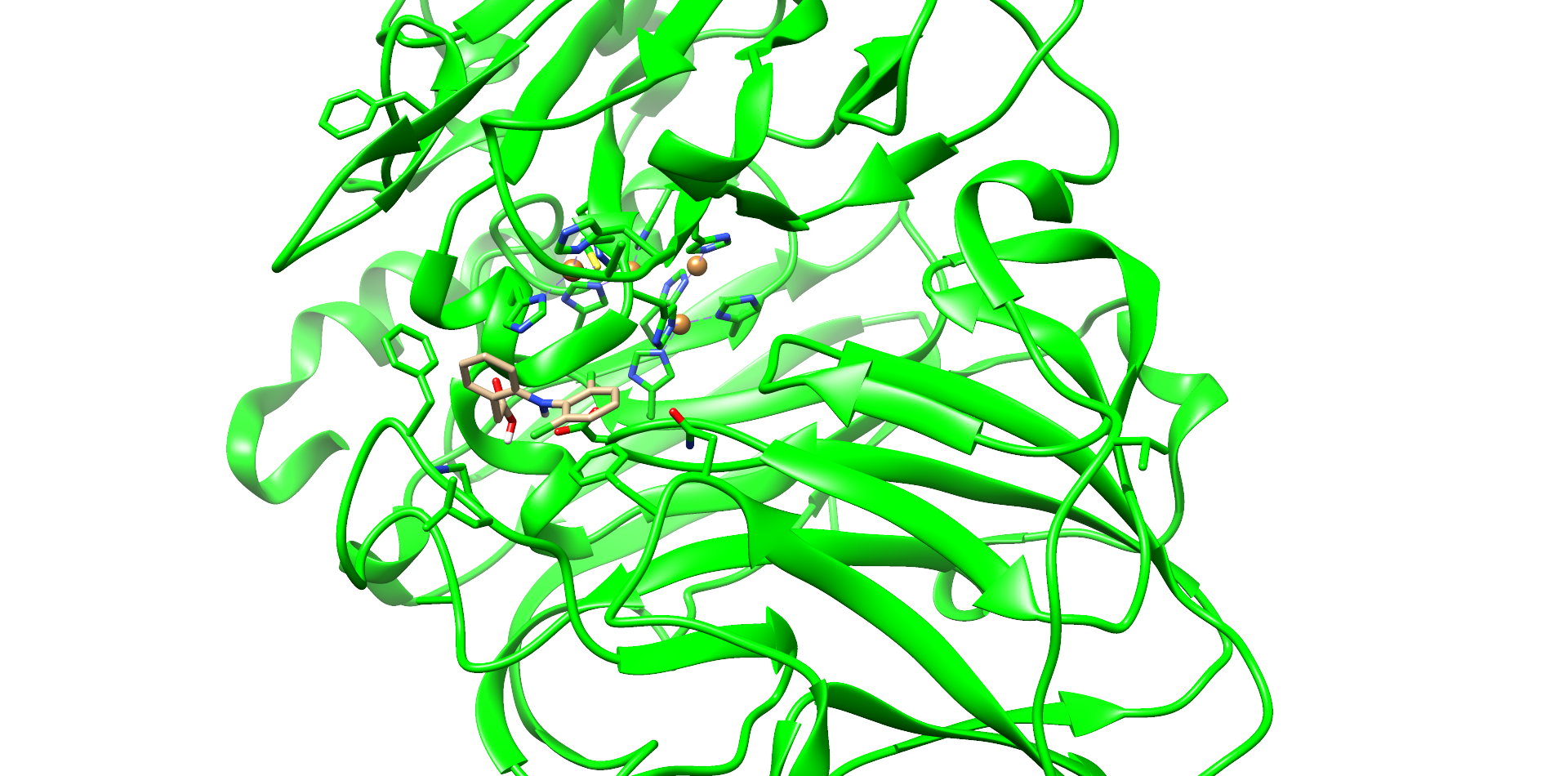
| + | One of these diclofenac structures in CotA is shown in Figure 5. Interestingly, diclofenac was positioned here in a <b>binding channel like structure</b> and not directly near the active copper atom within the active site. This binding channel structure is <b>flanked by two flexible loops</b> of CotA. These loops could undergo a conformational change due to the binding of the substrate, which could hold the substrate in place or even bring it closer to the active site. The <b>most dominant interaction</b> in this model were with the residue’s <b>threonine262</b> and <b>threonine418</b>. The threonine residues may interact with the secondary nitrogen and the carboxy group of diclofenac.<br> | ||
| + | </div> | ||
| + | <br> | ||
| + | |||
| + | <div style=" display: flex; | ||
| + | justify-content: center;"> | ||
| + | |||
| + | |||
| + | <figure class="figure"> | ||
| + | <img src="https://2020.igem.org/wiki/images/f/f5/T--TU_Darmstadt--CotA_1GSK_%2BDCF_Ribbon_01.png" width = "120%" alt="figure"> | ||
| + | |||
| + | </figure> | ||
| + | |||
| + | <figure class="figure"> | ||
| + | <img src="https://2020.igem.org/wiki/images/5/53/T--TU_Darmstadt--CotA_1GSK_DCF_Surface_01.png" alt="figure" width = "120%" > | ||
| + | |||
| + | </figure> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | <figcaption id="Figure5"><b>Figure 5: Diclofenac docking with CotA.</b> One of the best low energy interface score structures of CotA with diclofenac. Residues of the active site and in diclofenac binding involved residues are shown with sticks and colored heteroatoms. Diclofenac binds into a cavity and interacts with T262 and T418.</figcaption> | ||
| + | </div> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <div class="containertext"> | ||
| + | For comparison, the same docking procedure was performed with diclofenac and the laccase of T. versicolor. A structure with top scoring was also selected (Fig. 6). In this model diclofenac binds closer to the active site of the laccase of <i>T. Versicolor</i>. The substance also bound within a cavity of the enzyme and may interact by π-π-stacking with phenylalanine 163. Other interaction with the proline 163 and the carboxy group and aspartate 206 with the secondary nitrogen of diclofenac. The docking with the laccase from <i>T. Versicolor</i> showed more possible interactions and a better positioning of diclofenac near the active site. This assumption was supported by the interface and total score values of both docking experiments (table 2).<br> | ||
| + | </div> | ||
| + | <br> | ||
| + | |||
| + | <div style=" display: flex; | ||
| + | justify-content: center;"> | ||
| + | |||
| + | |||
| + | <figure class="figure"> | ||
| + | <img src="https://2020.igem.org/wiki/images/4/40/T--TU_Darmstadt--T.Vers_1GYC_DCF_Ribbon_01.png" class="figure-img img-fluid rounded" alt="T Versicolor Ribbon" width="120%"> | ||
| + | |||
| + | </figure> | ||
| + | |||
| + | <figure class="figure"> | ||
| + | <img src="https://2020.igem.org/wiki/images/a/af/T--TU_Darmstadt--T.Vers_1GYC_DCF_Surface_02.png" class="figure-img img-fluid rounded" alt="T Versicolor Surface" width="120%"> | ||
| + | |||
| + | </figure> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | <figcaption id="Figure5"><b>Figure 6: Diclofenac docking with the laccase from <i>T. Versicolor</i>.</b> One of the best low energy interface score structures of <i>T. Versicolor</i> laccase and diclofenac. Residues of the active site and in diclofenac binding involved residues are shown as sticks and with colored heteroatoms. Diclofenac binds into a cavity and interacts with P163, F265, D206.</figcaption> | ||
| + | </div> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <div class="containertext"> | ||
| + | The following table 2 also shows an overview of the different scoring values of the top scored structures from different substrates like <b>4-Nonylphenol, Ibuprofen and Carbamazepin</b> with CotA. The scoring values of all these substrates were relatively similar. Only the scoring values for the laccase from T. Versicolor were slightly better. But taking into account the positioning of the substance <i>T. Versicolor</i> is the superior laccase.<br> | ||
| + | </div> | ||
| + | <br> | ||
| + | |||
| + | <figcaption id="table2"><b>Table 2: Overview of the best interface and total score values from the docking experiments with CotA und <i>T. Versicolor</i>.</b> </figcaption> | ||
| + | <table class="tg"> | ||
| + | <thead> | ||
| + | <tr> | ||
| + | <th class="tg-7btt"> <br>Enzyme </th> | ||
| + | <th class="tg-l2oz"> <br>Substance </th> | ||
| + | <th class="tg-l2oz"> <br>Best Interface score </th> | ||
| + | <th class="tg-l2oz"> <br>Best Total score </th> | ||
| + | </tr> | ||
| + | </thead> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td class="tg-utjk" rowspan="4"> <br>CotA </td> | ||
| + | <td class="tg-lqy6"> <br>4-Nonylphenol </td> | ||
| + | <td class="tg-baqh"> <br>-11.014 </td> | ||
| + | <td class="tg-baqh"> <br>-1232.434 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td class="tg-lqy6"> <br>Ibuprofen </td> | ||
| + | <td class="tg-baqh"> <br>-12.170 </td> | ||
| + | <td class="tg-baqh"> <br>-1232.533 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td class="tg-lqy6"> <br>Carbamazepin </td> | ||
| + | <td class="tg-baqh"> <br>-12.633 </td> | ||
| + | <td class="tg-baqh"> <br>-1230.243 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td class="tg-lqy6"> <br>Diclofenac </td> | ||
| + | <td class="tg-baqh"> <br>-12.820 </td> | ||
| + | <td class="tg-baqh"> <br>-1230.488 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td class="tg-c85s"> <br>T. Versicolor </td> | ||
| + | <td class="tg-lqy6"> <br>Diclofenac </td> | ||
| + | <td class="tg-baqh"> <br>-13.266 </td> | ||
| + | <td class="tg-baqh"> <br>-1528.821 </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <div class="containertext"> | ||
| + | It is also confirmed in our docking experiments, that <i>T. Versicolor</i> binds diclofenac more optimal and thus may naturally degrade this substrate better. For a reliable confirmation of our results, we could further perform a <b>Quantum Mechanical (QM) docking procedure</b><sup id="cite_ref-18"><a href="#cite_note-18">[18]</sup></a>. But possible <b>starting points for modifications could be shown</b>. Consequently, we could start modifying our bacterial laccase by generating <b>mutants with the aspartate and phenylalanine residues</b> in similar loop region to the laccase of <i>T. Versicolor</i>. This procedure could possibly improve the binding affinity of diclofenac to CotA und therefore improve the ability of degrading this toxic substance by enzymatic oxidation. The many advantages of bacterial enzymes outweigh the use of fungal enzymes. For example, bacterial enzymes are a lot easier to produce in a greater scale, do not have complex glycosylation patterns and are prone to modifications. For this reason, it is worth pursuing these approaches. | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | <!-- Add more about the biology of this part here | ||
| + | ===Usage and Biology=== | ||
| + | |||
Revision as of 18:01, 25 October 2020
Laccase CotA
Profile
| Name | CotA-Laccase (Spore coat protein A) |
| Base pairs | 1542 |
| Molecular weight | 59.23 kDa |
| Origin | Bacillus subtilis, synthetic |
| Properties | CotA laccase from B. subtilis is a copper dependant oxireductase with a broad substrate variety of polyphenolic substrates such as diclofenac and ABTS and possess a T1 copper for primary electron transfer to the substrate and a T2/3 Cu-cluster. |
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
CotA Docking
At first we did the ligand docking of diclofenac with the laccase CotA. Preparations of the ligand and protein were done as explained above. For realization of the docking process we needed a working Rosetta xml script for our purpose. The script we worked out is shown in our Rosetta Guide. With this script, adjusted over many trials, we docked many thousand structures. As output we chose the Rosetta total score and the Rosetta interface score. We plotted these scores against the root-mean-square deviation of atomic positions (RMSD) from the ligand to allow a visible evaluation of the docked structures respectively poses[9]. The formula for calculation of the RMSD is shown below (formula 1).
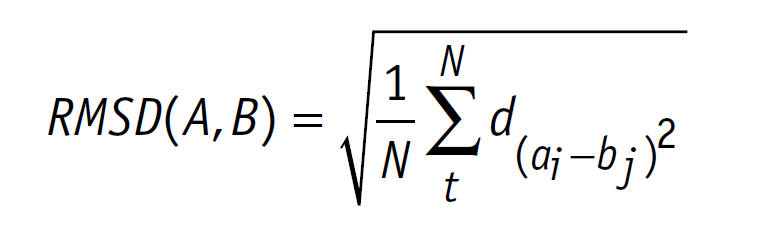
An interface score against RMSD plot from Diclofenac with CotA is shown in figure 4. Based on these plots it was easier to identify low energy structures with good interactions with the enzyme. Ideally the data points of these plots should form a funnel like plot. The structures on the closing end are especially good structures. These structures are highlighted with by a red square in figure 4.

One of these diclofenac structures in CotA is shown in Figure 5. Interestingly, diclofenac was positioned here in a binding channel like structure and not directly near the active copper atom within the active site. This binding channel structure is flanked by two flexible loops of CotA. These loops could undergo a conformational change due to the binding of the substrate, which could hold the substrate in place or even bring it closer to the active site. The most dominant interaction in this model were with the residue’s threonine262 and threonine418. The threonine residues may interact with the secondary nitrogen and the carboxy group of diclofenac.


For comparison, the same docking procedure was performed with diclofenac and the laccase of T. versicolor. A structure with top scoring was also selected (Fig. 6). In this model diclofenac binds closer to the active site of the laccase of T. Versicolor. The substance also bound within a cavity of the enzyme and may interact by π-π-stacking with phenylalanine 163. Other interaction with the proline 163 and the carboxy group and aspartate 206 with the secondary nitrogen of diclofenac. The docking with the laccase from T. Versicolor showed more possible interactions and a better positioning of diclofenac near the active site. This assumption was supported by the interface and total score values of both docking experiments (table 2).

The following table 2 also shows an overview of the different scoring values of the top scored structures from different substrates like 4-Nonylphenol, Ibuprofen and Carbamazepin with CotA. The scoring values of all these substrates were relatively similar. Only the scoring values for the laccase from T. Versicolor were slightly better. But taking into account the positioning of the substance T. Versicolor is the superior laccase.
| Enzyme |
Substance |
Best Interface score |
Best Total score |
|---|---|---|---|
| CotA |
4-Nonylphenol |
-11.014 |
-1232.434 |
| Ibuprofen |
-12.170 |
-1232.533 |
|
| Carbamazepin |
-12.633 |
-1230.243 |
|
| Diclofenac |
-12.820 |
-1230.488 |
|
| T. Versicolor |
Diclofenac |
-13.266 |
-1528.821 |
It is also confirmed in our docking experiments, that T. Versicolor binds diclofenac more optimal and thus may naturally degrade this substrate better. For a reliable confirmation of our results, we could further perform a Quantum Mechanical (QM) docking procedure[18]. But possible starting points for modifications could be shown. Consequently, we could start modifying our bacterial laccase by generating mutants with the aspartate and phenylalanine residues in similar loop region to the laccase of T. Versicolor. This procedure could possibly improve the binding affinity of diclofenac to CotA und therefore improve the ability of degrading this toxic substance by enzymatic oxidation. The many advantages of bacterial enzymes outweigh the use of fungal enzymes. For example, bacterial enzymes are a lot easier to produce in a greater scale, do not have complex glycosylation patterns and are prone to modifications. For this reason, it is worth pursuing these approaches.
