Difference between revisions of "Part:BBa K3202029"
Katherine Z4 (Talk | contribs) |
Katherine Z4 (Talk | contribs) |
||
| Line 12: | Line 12: | ||
As the result shows, both system exhibit relative high level of fluorescence compared to our backbone design( BN006) which suggest the two inducible transcription system function properly. | As the result shows, both system exhibit relative high level of fluorescence compared to our backbone design( BN006) which suggest the two inducible transcription system function properly. | ||
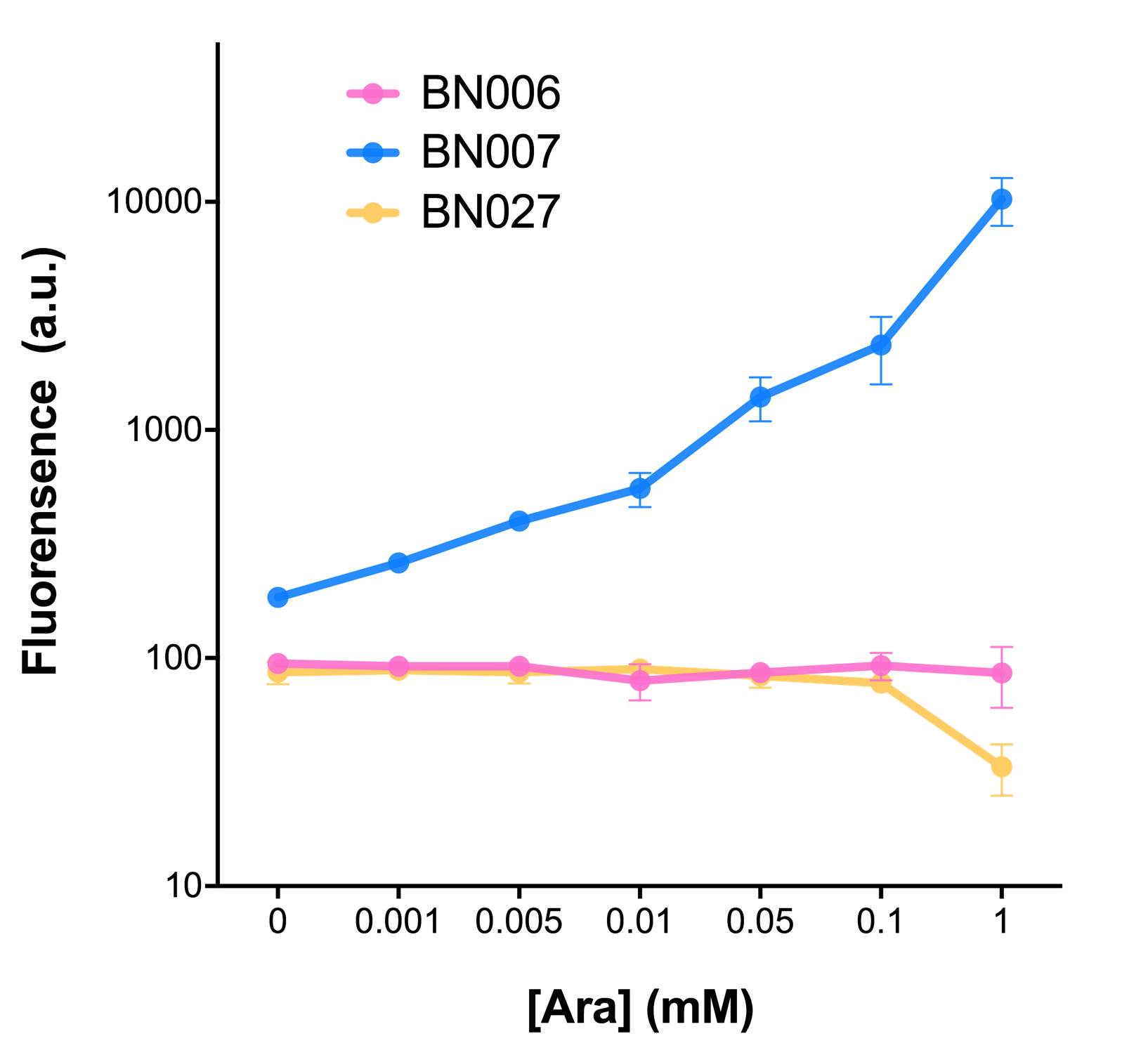
| − | [[File:T--BHSF_ND--Bistable_Experiment_5.png|500px|thumb| | + | [[File:T--BHSF_ND--Bistable_Experiment_5.png|500px|thumb|middle|alt=comparison between BN006 and BN007.|Figure 1: pBAD(BN007) induction curve at different concentrations after 4 hours compared to backbone.(BN006)]] |
Revision as of 10:45, 20 October 2019
Pc-Xyls
This composite part is designed as a genetic circuit that is used as a control group of our later experiment.
This acts as a control group in our experiment. It is compared to BBa_K3202030 and BBa_K3202031.
In our experiment of leakage testing, we used low-copying plasmid backbone PSB4K5, with a PSC101 origin of replication.We mainly focused on two types of inducible transcriptional promoter—XylS and pBAD. The XylS protein is the positive transcription regulator of the TOL plasmid meta-cleavage pathway operon Pm. XylS belongs to the AraC family of transcriptional regulators and exhibits an N-terminal domain involved in effector recognition, and a C-terminal domain. 【2】The activator XylS is usually is usually activated by the benzoate and its derivatives (3Mbz in our design). The pBAD promoter holds great potential to control the expression of genes that require tight regulation, as it is capable of both an induction and repression. The pBAD promoter exhibits an almost all-or-none behavior upon induction with arabinose when located on a high copy vector, but allows for gradual induction when cloned into a low copy vector.【3】 Based on these research findings, a low copy vector was used to investigate the ability to inhibit gene expression subsequent to induction of pBAD. As the result shows, both system exhibit relative high level of fluorescence compared to our backbone design( BN006) which suggest the two inducible transcription system function properly.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 93
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 815
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 278