Difference between revisions of "Part:BBa K3016600"
| Line 3: | Line 3: | ||
<partinfo>BBa_K3016600 short</partinfo> | <partinfo>BBa_K3016600 short</partinfo> | ||
| − | + | This part contains the CDS of YGFP, a slow bleaching variant of GFP. | |
| + | |||
| + | |||
| + | ==Characterization== | ||
| + | |||
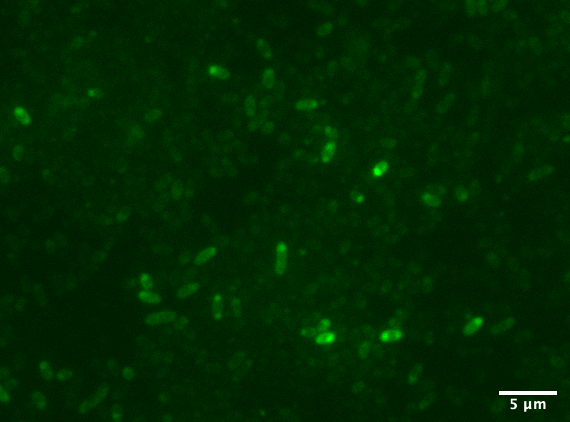
| + | Aalto-Helsinki characterized this part by fusing it with <i>Vibrio natriegens'</i> TorA Tat signal peptide ([https://parts.igem.org/Part:BBa_K3016100 BBa_K3016100]), creating a composite TorA-YGFP part ([https://parts.igem.org/Part:BBa_K3016200 BBa_K3016200]), to test its functionality in protein localisation studies in <i>V. natriegens</i> and <i>Escherichia coli</i> DH5a. | ||
| + | |||
| + | |||
| + | [[Image:T--Aalto-Helsinki--TorA-YGFP_natriegens.png|thumb|530px|center|<font size="1">Vibrio natriegens cells expressing TorA-YGFP</font>]] | ||
| + | |||
| + | |||
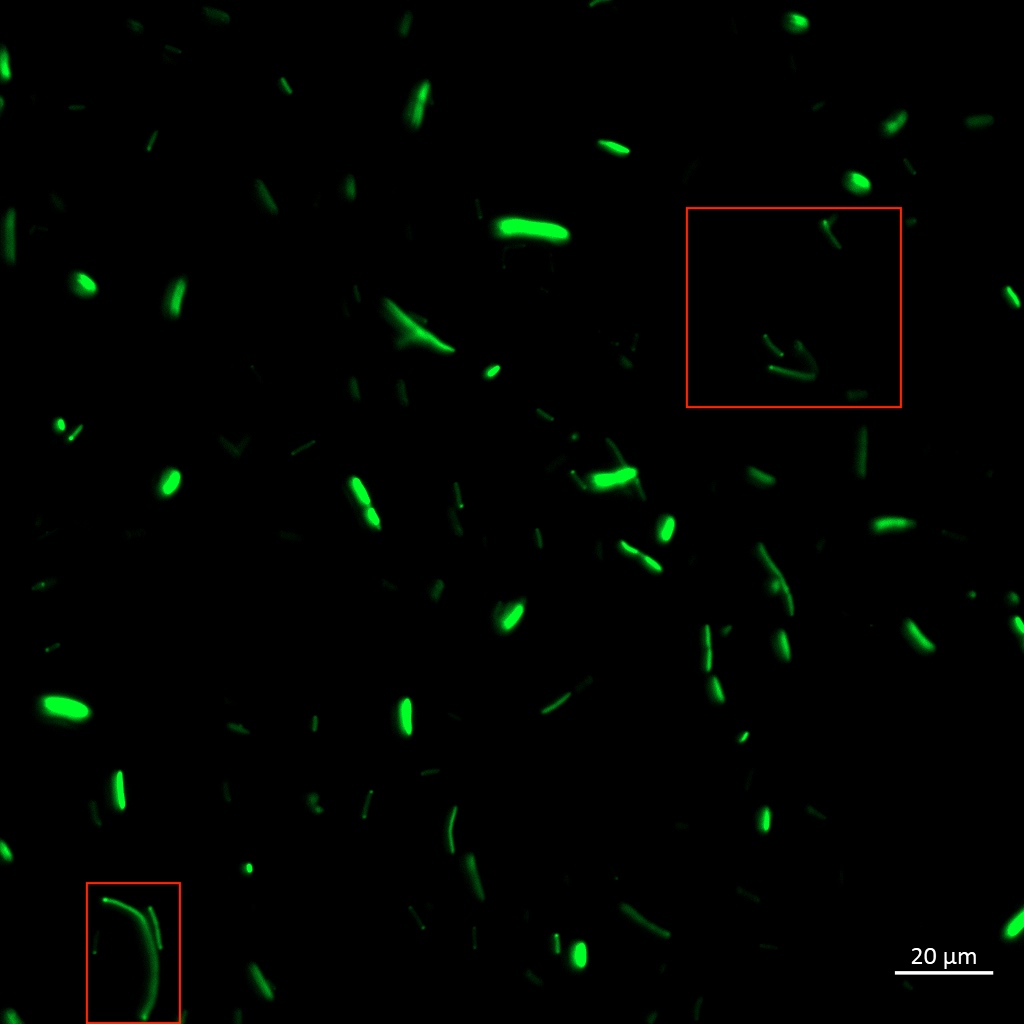
| + | [[Image:T--Aalto-Helsinki--TorA-YGFP_coli.jpg|thumb|530px|center|<font size="1"><i>Escherichia coli</i> DH5a cells expressing TorA-YGFP</font>]] | ||
| + | |||
| + | |||
| + | In the images above we see <i>Vibrio natriegens</i> and <i>Escherichia coli</i> DH5a cells expressing TorA-YGFP, which clearly fluoresces in both bacteria. Note the polar localisation of fluorescence. This can be a sign of periplasmic localisation of YGFP under osmotic pressure (Sochacki <i>et al.</i>, 2011) or inclusion body formation (Jong <i>et al.</i>, 2017). | ||
| + | |||
| + | |||
| + | To be certain of successful periplasmic translocation, a cell fractionation experiment was performed on <i>Vibrio natriegens</i>. The periplasmic fraction of TorA-YGFP expressing cells was extracted ([ protocol]) and placed under UV light, image below. The presence of YGFP in the periplasmic fractions can be seen clearly. | ||
| + | |||
| + | |||
| + | [[Image:T--Aalto-Helsinki--TorA-YGFP_natriegens_periplasmic_fraction.jpg|thumb|530px|center|<font size="1">Periplasmic fractions of Vibrio natriegens cells expressing TorA-YGFP </font>]] | ||
| + | |||
| + | |||
| + | These results indicate that the YGFP protein is active in both <i>Vibrio natriegens'</i> and <i>Escherichia coli</i> DH5a, and that it can successfully translocate into the periplasm of <i>Vibrio natriegens</i>, and possibly even of <i>Escherichia coli</i>, via the Tat pathway. | ||
| − | |||
| − | |||
<!-- --> | <!-- --> | ||
| Line 12: | Line 33: | ||
<partinfo>BBa_K3016600 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3016600 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | ==References:== | ||
| + | |||
| + | Alanen, H. I., Walker, K. L., Suberbie, M. L. V., Matos, C. F., Bönisch, S., Freedman, R. B., ... & Robinson, C. (2015). Efficient export of human growth hormone, interferon α2b and antibody fragments to the periplasm by the Escherichia coli Tat pathway in the absence of prior disulfide bond formation. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1853(3), 756-763. | ||
| + | |||
| + | Jong, W. S., Vikström, D., Houben, D., de Gier, J. W., & Luirink, J. (2017). Application of an E. coli signal sequence as a versatile inclusion body tag. Microbial cell factories, 16(1), 50. | ||
| + | |||
| + | Sochacki, K. A., Shkel, I. A., Record, M. T., & Weisshaar, J. C. (2011). Protein diffusion in the periplasm of E. coli under osmotic stress. Biophysical journal, 100(1), 22-31. | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Revision as of 11:36, 21 October 2019
YGFP, a slow bleaching spectral variant of GFP
This part contains the CDS of YGFP, a slow bleaching variant of GFP.
Characterization
Aalto-Helsinki characterized this part by fusing it with Vibrio natriegens' TorA Tat signal peptide (BBa_K3016100), creating a composite TorA-YGFP part (BBa_K3016200), to test its functionality in protein localisation studies in V. natriegens and Escherichia coli DH5a.
In the images above we see Vibrio natriegens and Escherichia coli DH5a cells expressing TorA-YGFP, which clearly fluoresces in both bacteria. Note the polar localisation of fluorescence. This can be a sign of periplasmic localisation of YGFP under osmotic pressure (Sochacki et al., 2011) or inclusion body formation (Jong et al., 2017).
To be certain of successful periplasmic translocation, a cell fractionation experiment was performed on Vibrio natriegens. The periplasmic fraction of TorA-YGFP expressing cells was extracted ([ protocol]) and placed under UV light, image below. The presence of YGFP in the periplasmic fractions can be seen clearly.
These results indicate that the YGFP protein is active in both Vibrio natriegens' and Escherichia coli DH5a, and that it can successfully translocate into the periplasm of Vibrio natriegens, and possibly even of Escherichia coli, via the Tat pathway.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 679
- 1000COMPATIBLE WITH RFC[1000]
References:
Alanen, H. I., Walker, K. L., Suberbie, M. L. V., Matos, C. F., Bönisch, S., Freedman, R. B., ... & Robinson, C. (2015). Efficient export of human growth hormone, interferon α2b and antibody fragments to the periplasm by the Escherichia coli Tat pathway in the absence of prior disulfide bond formation. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1853(3), 756-763.
Jong, W. S., Vikström, D., Houben, D., de Gier, J. W., & Luirink, J. (2017). Application of an E. coli signal sequence as a versatile inclusion body tag. Microbial cell factories, 16(1), 50.
Sochacki, K. A., Shkel, I. A., Record, M. T., & Weisshaar, J. C. (2011). Protein diffusion in the periplasm of E. coli under osmotic stress. Biophysical journal, 100(1), 22-31.