Difference between revisions of "Part:BBa K3013003"
m (→Usage and Biology) |
m (→Usage and Biology) |
||
| Line 14: | Line 14: | ||
This part and <partinfo>BBa_K1094401</partinfo> were cloned into the expression vector pET28a(+) via restriction and ligation. The expression vector contained a T7 promoter (pT7) and a T7 terminator. As growth of Magnetospirillum sp. and R. rubrum magneticum is low, E. coli was chosen for demonstration purposes. So, both constructs were transformed into chemically competent E. coli BL21. Correct clones were selected via colony PCR. 10 μL of the PCR product was loaded on a 1,2 % agarose gel (gel was stained by RotiSafe). A picture of the gel is shown in figure 1.<br> | This part and <partinfo>BBa_K1094401</partinfo> were cloned into the expression vector pET28a(+) via restriction and ligation. The expression vector contained a T7 promoter (pT7) and a T7 terminator. As growth of Magnetospirillum sp. and R. rubrum magneticum is low, E. coli was chosen for demonstration purposes. So, both constructs were transformed into chemically competent E. coli BL21. Correct clones were selected via colony PCR. 10 μL of the PCR product was loaded on a 1,2 % agarose gel (gel was stained by RotiSafe). A picture of the gel is shown in figure 1.<br> | ||
<br> | <br> | ||
| − | <gallery heights= | + | <gallery heights=200px widths=500> |
Image: Validation gel gold.png| '''Figure 1:''' ''selection of correct clones via colony PCR. 10 μL of PCR product was loaded on 1,2 % agarose gel (120 V, 300 mA, 50 W, 25 min). 1 kb+ ladder was used in the outer lines. PCR was made of 9 colonies after transformation and selection on LB-Kan plates of cells that should contain the mamC-eGFP and the mamC-FbFP BS2 construct. Expected bands for mamC-eGFp are 1200 bp and for mamC-FbFP BS2 are 800.'' | Image: Validation gel gold.png| '''Figure 1:''' ''selection of correct clones via colony PCR. 10 μL of PCR product was loaded on 1,2 % agarose gel (120 V, 300 mA, 50 W, 25 min). 1 kb+ ladder was used in the outer lines. PCR was made of 9 colonies after transformation and selection on LB-Kan plates of cells that should contain the mamC-eGFP and the mamC-FbFP BS2 construct. Expected bands for mamC-eGFp are 1200 bp and for mamC-FbFP BS2 are 800.'' | ||
</gallery> | </gallery> | ||
Revision as of 15:46, 20 October 2019
mamC-FbFP BS2 fusion
This part is designed as an improvement of part BBa_K1094401.
This fusion protein consists of mamC, a glycine linker and a fluorescent protein. MamC is suggested to be located in the magnetosome membrane, which are produced by magnetotactic bacteria. To report expression of mamC FbFP BS2 is used.
Magnetotactic bacteria produce magnetosomes under microaerobic conditions [Heyen U, Schüler D (2003) Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl Microbiol Biotechnol 61:536–544]. As it requires elaborate equipment to setup such conditions, a simpler approach was tested by team Aachen 2019 which was cultivation of Magnetospirillum sp. and Rhodospirillum rubrum magneticum in closed flasks. Formation of magnetosomes was confirmed via transmission electron microscopy for R. rubrum magneticum and by formation of culture in a magnetic field for Magnetospirillum gryphiswaldense. Dissolved oxygen concentrations after days of cultivation tend towards zero. As maturation of GFP is dependent to oxygen, problems with reporting mamC expression levels under microaerobic or anaerobic conditions could arise [Tsien, R. Y. The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 (1998)]. Therefore, a more reliable reporter for microaerobic conditions with unknown oxygen concentrations is needed. Such a reporter is FbFP BS2 (BBa_K3013002). As its fluorophore formation is FMN-based (flavin mono nucleotide), it is not dependent to oxygen. We chose this fluorescent protein as it is also smaller than eGFP (eGFP: ~720 bp, FbFP BS2: 408 bp) and expression of the reporter should be as stressfree as possible for the host strain as well as the size of bigger fusion proteins like mamC-FbFP BS2-LCI (BBa_K3013000) should stay as small as possible for better expression results.
Usage and Biology
This part and BBa_K1094401 were cloned into the expression vector pET28a(+) via restriction and ligation. The expression vector contained a T7 promoter (pT7) and a T7 terminator. As growth of Magnetospirillum sp. and R. rubrum magneticum is low, E. coli was chosen for demonstration purposes. So, both constructs were transformed into chemically competent E. coli BL21. Correct clones were selected via colony PCR. 10 μL of the PCR product was loaded on a 1,2 % agarose gel (gel was stained by RotiSafe). A picture of the gel is shown in figure 1.
Figure 1: selection of correct clones via colony PCR. 10 μL of PCR product was loaded on 1,2 % agarose gel (120 V, 300 mA, 50 W, 25 min). 1 kb+ ladder was used in the outer lines. PCR was made of 9 colonies after transformation and selection on LB-Kan plates of cells that should contain the mamC-eGFP and the mamC-FbFP BS2 construct. Expected bands for mamC-eGFp are 1200 bp and for mamC-FbFP BS2 are 800.
As shown in figure 1, insertion of mamC-constructs was successful.
As expression with anaerobic conditions was not possible in our lab and data for eGFP expression in anaerobic conditions is available, we decided to compare both reporters in aerobic conditions.
Main cultures were inoculated from precultures and induced at OD(600) = 0,7 with 0,1 mM IPTG (final concentration). Proteins were expressed at 30 °C for 16 h on shaker (250 rpm).
Cells were observed via microscopy at 100x magnification. Pictures were taken in brightfield and with filter setting for eGFP (Wavelength could not be set manually, so eGFP filter setup was used for FbFP BS2 too). Resulting pictures are shown in figure 2.
Figure 2: microscopy of cells 16 h after induction of protein expression at 30 °C with IPTG (0,1 mM final concentration) with 100x magnification. Pictures A and C are taken in brightfield while pictures B and D are taken with filter setup for eGFP. Intensity of light are the same for pictures C and D. Pictures A and B: mamC-eGFP expression in E. coli BL21, C and D: mamC-FbFP BS2 expression in E. coli BL21.
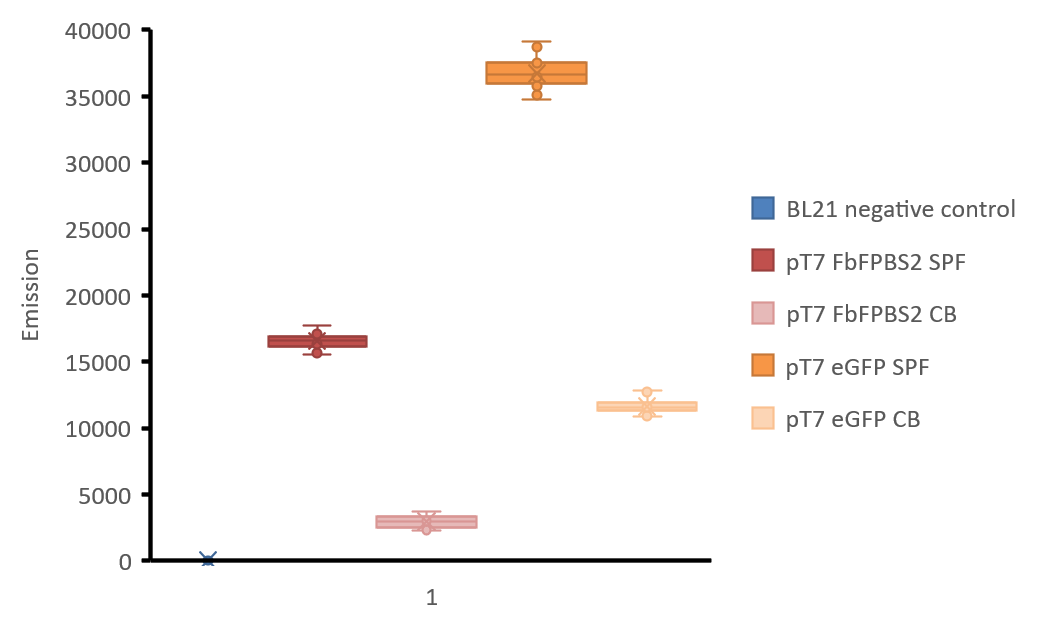
Figure 3: Boxplot-diagram of fluorescence intensities. All points are shown. Measurement was made with Tecan Infinite M1000 reader in a black 96 well plate. 4 points of measurement were set in each well (number of flashes: 50, flash frequency: 400 Hz, integration time: 20 μs, lag time: 0 μs, settle time: 10 μs. Z-position was calculated from wells), gain was manually set to 98 after calculation of the optimal gain in one well containing the soluble protein fraction of mamC-eGFP. excitation: 450 nm and emission: 495 nm for mamC-FbFP BS2 and with excitation: 485 nm, emission: 535 nm for mamC-eGFP. BL21 negative control: soluble protein fraction of E. coli BL21 without any expression vector, soluble protein fractions of E. coli BL21 are indicated with “SPF” and cell broth is indicated with “CB”.
As shown in figure 3, the fluorescence of eGFP in aerobic conditions is higher than FbFP BS2. Fluorescence of cultures is always lower than the fluorescence of the purified protein fractions, but still the fluorescence signal of FbFP BS2 in E. coli culture is high enough for good detection. We did not express mamC-eGFP in anaerobic conditions but as data has shown, its fluorescence will not be present.
As FbFP BS2 maintained fluorescence in the fusion of mamC-FbFP BS2 and a good fluorescence signal was archived, we conclude that mamC-FbFP BS2 will be more reliable for in vivo detection of mamC expression levels in Magnetospirillum sp. or R. rubrum magneticum.
In addition, FbFP BS2 could also be used as a reporter in other applications and in vitro detection systems under anaerobic conditions.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 498
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 169
Illegal NgoMIV site found at 253
Illegal AgeI site found at 666 - 1000COMPATIBLE WITH RFC[1000]