Difference between revisions of "Part:BBa K2985004"
| Line 11: | Line 11: | ||
In order to overexpress the gene in Bacillus subtilis, we designed a unique fragment, which can integrate the target gene into the genome of Bacillus subtilis. | In order to overexpress the gene in Bacillus subtilis, we designed a unique fragment, which can integrate the target gene into the genome of Bacillus subtilis. | ||
| − | [[File:Jiangnan-China ycxA-figure 1. | + | [[File:Jiangnan-China ycxA-figure 1.jpeg]]<br> |
'''Figure 1: Upstream homologous arm (742bp) + SPC resistance gene (1104bp) + P43 promoter (678 / 56bp) + ycxA (1227bp) + downstream homologous arm(646bp)''' | '''Figure 1: Upstream homologous arm (742bp) + SPC resistance gene (1104bp) + P43 promoter (678 / 56bp) + ycxA (1227bp) + downstream homologous arm(646bp)''' | ||
| Line 18: | Line 18: | ||
In order to determine the length of the sequence, we performed partial PCR experiments and obtained gel map to determine whether the length of the sequences was correct or not. | In order to determine the length of the sequence, we performed partial PCR experiments and obtained gel map to determine whether the length of the sequences was correct or not. | ||
| − | [[File:Jiangnan-China ycxA-figure 2. | + | [[File:Jiangnan-China ycxA-figure 2.jpeg]]<br> |
'''Figure 2 0.8% agarose gel electrophoresis and PCR validation. The product has a signal band at 1104bp, consistent with the target length.''' | '''Figure 2 0.8% agarose gel electrophoresis and PCR validation. The product has a signal band at 1104bp, consistent with the target length.''' | ||
| − | [[File:Jiangnan-China ycxA-figure 3. | + | [[File:Jiangnan-China ycxA-figure 3.jpeg]]<br> |
'''Figure 3 0.8% agarose gel electrophoresis and PCR validation. The product has a signal band at 1227bp, consistent with the target length.''' | '''Figure 3 0.8% agarose gel electrophoresis and PCR validation. The product has a signal band at 1227bp, consistent with the target length.''' | ||
| Line 28: | Line 28: | ||
First, co culture with wild-type Escherichia coli. After a total of 12 hours of culture, we coated plate culture, and obtained the morphological differences of E.coli, in which small colonies represent E.coli and large colonies represent Bacillus subtilis. In the same condition, the survival rate of E.coli in co culture with Bacillus subtilis after enhanced efflux gene was lower, except for the difference of Bacillus subtilis. We speculated that the growth of E.coli was inhibited by the increase of surfactin production.<br> | First, co culture with wild-type Escherichia coli. After a total of 12 hours of culture, we coated plate culture, and obtained the morphological differences of E.coli, in which small colonies represent E.coli and large colonies represent Bacillus subtilis. In the same condition, the survival rate of E.coli in co culture with Bacillus subtilis after enhanced efflux gene was lower, except for the difference of Bacillus subtilis. We speculated that the growth of E.coli was inhibited by the increase of surfactin production.<br> | ||
| − | [[File:Jiangnan-China ycxA-figure 4. | + | [[File:Jiangnan-China ycxA-figure 4.jpeg]]<br> |
'''Figure 4 Co-culture of wild type bacillus subtilis and Escherichia coli'''<br> | '''Figure 4 Co-culture of wild type bacillus subtilis and Escherichia coli'''<br> | ||
| − | [[File:Jiangnan-China ycxA-figure 5. | + | [[File:Jiangnan-China ycxA-figure 5.jpeg]]<br> |
'''Figure 5 Co-culture of Bacillus subtilis and Escherichia coli after transformation'''<br> | '''Figure 5 Co-culture of Bacillus subtilis and Escherichia coli after transformation'''<br> | ||
In addition, we also did the blood agar plate test, but the result of hemolysis circle is not very obvious. | In addition, we also did the blood agar plate test, but the result of hemolysis circle is not very obvious. | ||
| − | [[File:Jiangnan-China ycxA-figure 6. | + | [[File:Jiangnan-China ycxA-figure 6.jpeg]]<br> |
'''Figure 6 Elatively small colonies producing surfactin before transformation'''<br> | '''Figure 6 Elatively small colonies producing surfactin before transformation'''<br> | ||
| − | [[File:Jiangnan-China ycxA-figure 7. | + | [[File:Jiangnan-China ycxA-figure 7.jpeg]]<br> |
'''Figure 7 The colony of bacillus subtilis after transformation is relatively large and the hemolysis circle is larger.'''<br> | '''Figure 7 The colony of bacillus subtilis after transformation is relatively large and the hemolysis circle is larger.'''<br> | ||
Revision as of 03:30, 20 October 2019
ycxA->ycxA gene encodes a specific carrier protein. In the process of cell growth, YcxA protein effe
The ycxA gene is located in surfactin synthase gene cluster downstream of srfA operon. It encodes proteins homologous to members of the main promoter superfamily. It is mainly related to active transport of lipopeptides. The length of ycxa coding region is 1227 bp. According to Xu, ycxA protein is mainly a transmembrane protein, which participates in the export of surfactin and trans-proton gradient transport mechanism. Overexpression of ycxA gene resulted in a marked increase in surfactin production. Meanwhile, hemolytic circles were observed in single cells cultured on agar plates.
According to the literature of Xu [1], Ycxa protein is mainly a transmembrane protein, which is involved in the export of surfacin and the transport mechanism of cross proton gradient. Under the overexpression of ycxa gene, the production of surfacin increased significantly. At the same time, the hemolysis circle of single cell was more obvious in the blood agar plate culture.
The position of ycxa gene is mainly in the middle of srfA and sfp, which is closely related to the efflux of surfacin. In order to enhance the expression of this gene and make surfacin efflux in bacteria, we overexpress this gene, reduce the intracellular accumulation of surfacin, and increase the yield of surfacin.
In order to overexpress the gene in Bacillus subtilis, we designed a unique fragment, which can integrate the target gene into the genome of Bacillus subtilis.
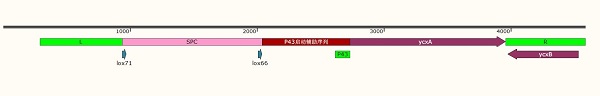
Figure 1: Upstream homologous arm (742bp) + SPC resistance gene (1104bp) + P43 promoter (678 / 56bp) + ycxA (1227bp) + downstream homologous arm(646bp)
We send the sequence information to biotechnology company, and synthesize the fragments we need.
In order to determine the length of the sequence, we performed partial PCR experiments and obtained gel map to determine whether the length of the sequences was correct or not.
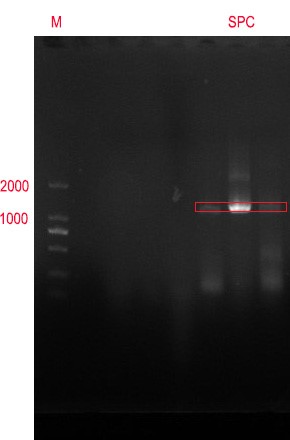
Figure 2 0.8% agarose gel electrophoresis and PCR validation. The product has a signal band at 1104bp, consistent with the target length.
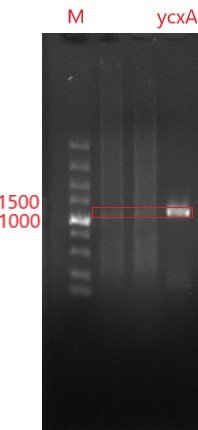
Figure 3 0.8% agarose gel electrophoresis and PCR validation. The product has a signal band at 1227bp, consistent with the target length.
After the successful transformation of ycxA gene, in order to study the specific value and function of modified components, we designed two kinds of validation experiments to distinguish the specific differences before and after the transformation.
First, co culture with wild-type Escherichia coli. After a total of 12 hours of culture, we coated plate culture, and obtained the morphological differences of E.coli, in which small colonies represent E.coli and large colonies represent Bacillus subtilis. In the same condition, the survival rate of E.coli in co culture with Bacillus subtilis after enhanced efflux gene was lower, except for the difference of Bacillus subtilis. We speculated that the growth of E.coli was inhibited by the increase of surfactin production.
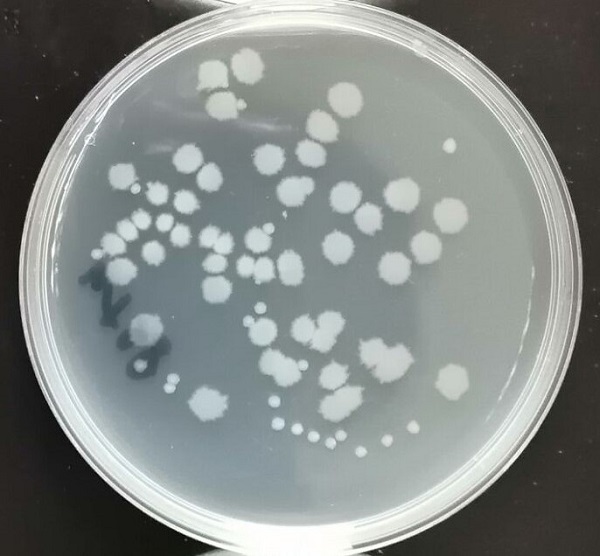
Figure 4 Co-culture of wild type bacillus subtilis and Escherichia coli
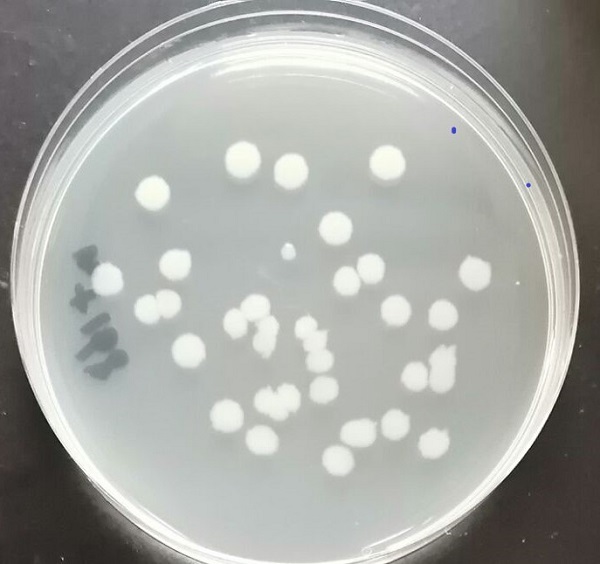
Figure 5 Co-culture of Bacillus subtilis and Escherichia coli after transformation
In addition, we also did the blood agar plate test, but the result of hemolysis circle is not very obvious.
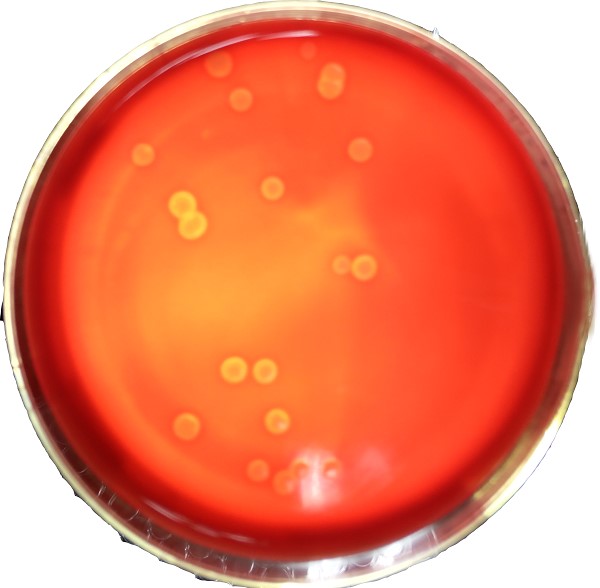
Figure 6 Elatively small colonies producing surfactin before transformation
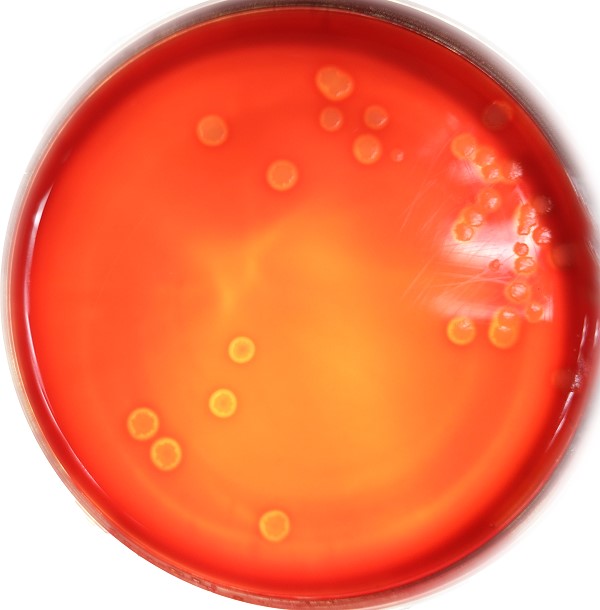
Figure 7 The colony of bacillus subtilis after transformation is relatively large and the hemolysis circle is larger.
In addition, we also carried out high-performance liquid phase analysis of the yield of surfactin, which also proved that the parts for Bacillus subtilis production of surfactin ability. However, the increase in the yield of surfactin does not show the transcriptional level and quantifiable effect of ycxA gene, so we have no further study.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 905
- 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 905
- 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 905
Illegal BglII site found at 816 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 905
- 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 905
Illegal NgoMIV site found at 484 - 1000COMPATIBLE WITH RFC[1000]
