Difference between revisions of "Part:BBa K2407012"
| Line 18: | Line 18: | ||
<partinfo>BBa_K2407012 parameters</partinfo> | <partinfo>BBa_K2407012 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | |||
| + | ===Team: Tianjin 2019 characterization=== | ||
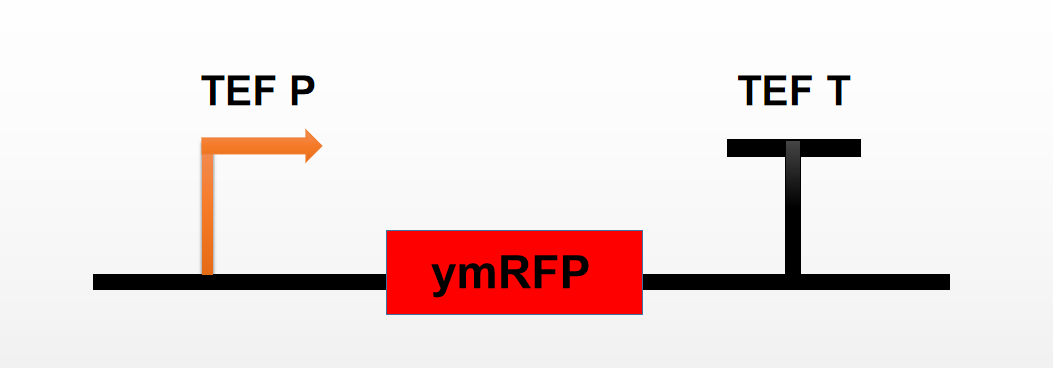
| + | Team:Tianjin 2019 contributed to the reporter's characterization by connecting it to the combined promoters TEF and testing their fluorescence intensity and detailed fluorescence excitation curves. | ||
| + | [[File:T--Tianjin--70121.png|500px|thumb|none|Figure 1. Characterization scheme used by Team Tianjin]] | ||
| + | We transformed the constructed fragment directly into Saccharomyces cerevisiae and cultured it overnight under normal conditions (YES solid medium, 30 ℃). On the next day, the colony was visible to the naked eye, and the appearance was reddish. | ||
| + | [[File:T--Tianjin--70122.png|500px|thumb|none|Fig 2.Cell morphology on medium]] | ||
| + | <br> | ||
| + | The experiment design<br> | ||
| + | 1. Fluorescence microplate reader measurement<br> | ||
| + | The red colonies on the medium were cultured alone overnight. Then take 200 μL of bacterial solution, centrifugate it for 2 min at 4000R, pour out the supernatant and then re spin it with 200 μ l of water, place it in the hole, take its culture medium as the control, and measure its equal bacterial amount fluorescence intensity under the emission / excitation of 584 / 607 (literature value).<br><br> | ||
| + | 2. Fluorescence curve determination<br> | ||
| + | We measured the specific fluorescence intensity at 470 ~ 600nm emission wavelength under the condition of fixed excitation wavelength (607nm), and obtained the following results:<br> | ||
| + | Then we measured the specific fluorescence intensity at 500 ~ 650nm under the condition of fixed emission wavelength (584nm), and obtained the following results: | ||
Revision as of 02:35, 22 October 2019
a modified yEmRFP for use in Yeast from mCherry mRFP
It is the RFP variant sequence that codon-optimized for the expression in Saccharomyces cerevisiae as yeast-enhanced mRFP (yEmRFP) and can combine fluorescence and a purple visible phenotype.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Team: Tianjin 2019 characterization
Team:Tianjin 2019 contributed to the reporter's characterization by connecting it to the combined promoters TEF and testing their fluorescence intensity and detailed fluorescence excitation curves.
We transformed the constructed fragment directly into Saccharomyces cerevisiae and cultured it overnight under normal conditions (YES solid medium, 30 ℃). On the next day, the colony was visible to the naked eye, and the appearance was reddish.
The experiment design
1. Fluorescence microplate reader measurement
The red colonies on the medium were cultured alone overnight. Then take 200 μL of bacterial solution, centrifugate it for 2 min at 4000R, pour out the supernatant and then re spin it with 200 μ l of water, place it in the hole, take its culture medium as the control, and measure its equal bacterial amount fluorescence intensity under the emission / excitation of 584 / 607 (literature value).
2. Fluorescence curve determination
We measured the specific fluorescence intensity at 470 ~ 600nm emission wavelength under the condition of fixed excitation wavelength (607nm), and obtained the following results:
Then we measured the specific fluorescence intensity at 500 ~ 650nm under the condition of fixed emission wavelength (584nm), and obtained the following results: