Difference between revisions of "Part:BBa K2201017"
| Line 4: | Line 4: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
This plasmid was designed as a backbone for the incorporation of fragments containing unnatural base pairs. | This plasmid was designed as a backbone for the incorporation of fragments containing unnatural base pairs. | ||
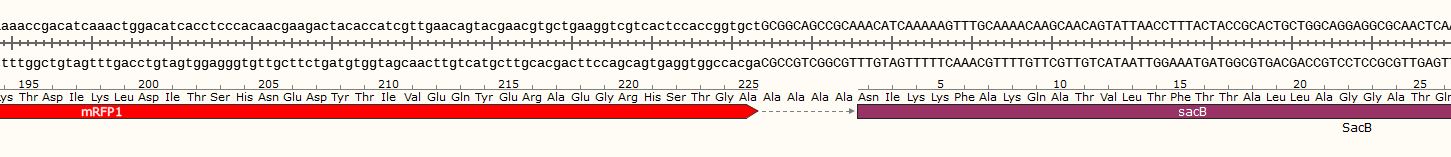
| − | Given that selection and screening of the transformants was not possible with the experimental design, a plasmid which would kill or inhibit growth of the wrong colonies was designed. The initial design focused on the use of CcdB, but given that <i>ccd</i>B cannot be handed in, levansucrase of <i>Bacillus subtilis</i>, encoded by <i>sac</i>B, was used instead. Therefore, a <i>mRFP-sac< | + | Given that selection and screening of the transformants was not possible with the experimental design, a plasmid which would kill or inhibit growth of the wrong colonies was designed. The initial design focused on the use of CcdB, but given that <i>ccd</i>B cannot be handed in, levansucrase of <i>Bacillus subtilis</i>, encoded by <i>sac</i>B, was used instead. Therefore, a <i>mRFP-sac</i>B fusion construct was constructed. BBa_J23100 was used as a promotor and BBa_B0034 as a strong RBS, followed by BBa_E1010. Between <i>mRFP</i> and <i>sac</i>B a linker consisting of four alanines was used. Given that this plasmid was designed to eventually carry an unnatural base pair, a low-copy plasmid was the better choice for a higher stability of the unnatural base pair. Therefore, the backbone pSB3C5 was used. |
Latest revision as of 12:50, 1 November 2017
Construct for the integration of unnatural base pairs
Usage and Biology
This plasmid was designed as a backbone for the incorporation of fragments containing unnatural base pairs. Given that selection and screening of the transformants was not possible with the experimental design, a plasmid which would kill or inhibit growth of the wrong colonies was designed. The initial design focused on the use of CcdB, but given that ccdB cannot be handed in, levansucrase of Bacillus subtilis, encoded by sacB, was used instead. Therefore, a mRFP-sacB fusion construct was constructed. BBa_J23100 was used as a promotor and BBa_B0034 as a strong RBS, followed by BBa_E1010. Between mRFP and sacB a linker consisting of four alanines was used. Given that this plasmid was designed to eventually carry an unnatural base pair, a low-copy plasmid was the better choice for a higher stability of the unnatural base pair. Therefore, the backbone pSB3C5 was used.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 616
Illegal AgeI site found at 728 - 1000COMPATIBLE WITH RFC[1000]
Contents
Plasmid Design
In the initial plasmid, mRFP and sacB are in-frame, meaning that cells turn red when incubated in standard media such as LB, but die or grow weakly when cultivated in media supplemented with sucrose. This is due to the toxic effect of levansucrase which unfolds when cells are cultivated in sucrose supplemented media. The linker between mRFP and sacB represents the insertion site for the fragment containing the unnatural base pair. This insertion, if successful, leads to a frameshift of the downstream located sacB, leading to loss of function. Therefore, cells can only survive in sucrose supplemented media if the fragment containing the unnatural base pair was successfully incorporated. This provides an easy screening method, since cells that turn red and survive in sucrose supplemented media most likely inserted the fragment containing the unnatural base pair into the plasmid. Having the coding sequences of both proteins in one frame has one big advantage: given that sacB is prone to loss-of-function mutations, contaminations can be easily distinguished by color. Furthermore, cells with mutations in the promotor region would also survive since they would not express sacB, but not turn red. Therefore, this two-layered system allows to visually check if a culture is contaminated, has a mutation in the promotor region or if the correct plasmid is present.

To test the effect of sucrose on growth of the cells, a cultivation was performed of E. coli BL21(DE3) harboring BBa_K2201017. The cultivations were carried out in a 12 well plate in 1 mL of LB media supplemented with different concentrations of sucrose. Three biological replicates were cultivated for each condition and three technical replicates taken for each measurement point.

The cultivations shown in figure (3) clearly show that sucrose has a significant effect on growth of strains carrying BBa_K2201017. A higher sucrose concentration leads to weaker growth, with strains cultivated in 20 % sucrose reaching only ~33 % of the OD600 of the reference strain cultivated in sucrose free media. The reference reached a final optical density of 3.233 ± 0.148, strains cultivated in media supplemented with 10 % sucrose 1.527 ± 0.055 and strains cultivated in 20 % sucrose reached a final optical density of 0.44 ± 0.05. Figure (4) shows a comparison of strains cultivated in media without sucrose and media supplemented with 10 % sucrose. A preculture of E. coli BL21(DE3) harboring BBa_K2201017 was prepared and cultivated overnight at 37 °C. 30 µl of this precultures were used and dropped onto LB agar plates either without or supplemented with 10 % sucrose. 50 µl of the same preculture were used to inoculate 3 mL of LB media without sucrose and 3 mL of LB media supplemented with 10 % sucrose.