Difference between revisions of "Part:BBa K1949103:Design"
| Line 20: | Line 20: | ||
-Plasmids | -Plasmids | ||
| − | + | <i>E. coli</i> A: Pcon - <i>rbs</i> - <i>gfp</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | |
| − | + | <i>E. coli</i> B: PBAD - <i>rbs - mazF - tt</i> - Pcon - <i>rbs - gfp</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | |
| − | =====Ⅱ.<i>mazEF</i> System Assay ~ | + | =====Ⅱ.<i>mazEF</i> System Assay ~Stop & GO~===== |
-Plasmids | -Plasmids | ||
| − | + | <i>E. coli</i> C: PBAD - <i>rbs</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | |
| − | + | <i>E. coli</i> A: Pcon - <i>rbs - gfp</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | |
| − | + | <i>E. coli</i> D: PBAD - <i>rbs - mazF - tt</i> - Pcon - <i>rbs - gfp</i> (pSB6A1), Plac - <i>rbs - mazE</i> (pSB3K3) | |
| − | + | <i>E. coli</i> B: PBAD - <i>rbs - mazF - tt</i> - Pcon - <i>rbs - gfp</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | |
| − | + | =====Ⅲ.<i>mazEF</i> System Assay ~Go & Stop~===== | |
| + | |||
| + | -Plasmids | ||
| + | |||
| + | <i>E. coli</i> C: PBAD - <i>rbs</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | ||
| + | |||
| + | <i>E. coli</i> A: Pcon - <i>rbs - gfp</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | ||
| + | |||
| + | <i>E. coli</i> G: PBAD - <i>rbs - mazF - tt</i> - Pcon - <i>rbs - gfp</i> (pSB6A1), Pcon - <i>rbs</i>(weak) - <i>mazE</i> (pSB3K3) | ||
| + | |||
| + | <i>E. coli</i> F: PBAD - <i>rbs - mazF - tt</i> - Pcon - <i>rbs - gfp</i> (pSB6A1), Pcon - <i>rbs - mazE</i> (pSB3K3) | ||
| + | |||
| + | <i>E. coli</i> E: PBAD - <i>rbs - mazF - tt</i> - Pcon - <i>rbs - gfp</i> (pSB6A1), vector (pSB3K3) | ||
| + | |||
| + | =====Ⅳ.Control of Cell Growth===== | ||
| + | |||
| + | -Plasmid | ||
| + | |||
| + | <i>E. coli</i> I: PBAD - <i>rbs</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | ||
| + | |||
| + | <i>E. coli</i> D: PBAD - <i>rbs - mazF</i> (pSB6A1), Plac - <i>rbs - mazE</i> (pSB3K3) | ||
| + | |||
| + | <i>E. coli</i> H: PBAD - <i>rbs - mazF</i>(pSB6A1), Plac - <i>rbs</i> (pSB3K3) | ||
====Assay protocol==== | ====Assay protocol==== | ||
| − | =====Ⅰ.Adjustment of | + | =====Ⅰ.Adjustment of <i>mazF</i> Expression===== |
| Line 58: | Line 80: | ||
5)Incubate with vigorous shaking for 24 h, and measure the turbidity and the RFU of GFP. | 5)Incubate with vigorous shaking for 24 h, and measure the turbidity and the RFU of GFP. | ||
| − | =====Ⅱ.<i>mazEF</i> System Assay ~ | + | =====Ⅱ.<i>mazEF</i> System Assay ~Stop & GO~===== |
======Pre-culture====== | ======Pre-culture====== | ||
1)Suspend colonies on a master plate into LB medium containing ampicillin (50 microg / mL) and kanamycin (50 microg / mL). | 1)Suspend colonies on a master plate into LB medium containing ampicillin (50 microg / mL) and kanamycin (50 microg / mL). | ||
| Line 67: | Line 89: | ||
1)Measure the turbidity of the pre-cultures. | 1)Measure the turbidity of the pre-cultures. | ||
| − | 2)Dilute the pre- cultures to 1 / 30 into LB medium containing 4 mL ampicillin and kanamycin. | + | 2)Dilute the pre-cultures to 1 / 30 into LB medium containing 4 mL ampicillin and kanamycin. |
| + | |||
| + | 3)Incubate with vigorous shaking so that turbidity becomes 0.03. | ||
| + | |||
| + | 4)Add arabinose so that the final concentration becomes 0.02%. | ||
| + | |||
| + | 5)Add IPTG until the concentration becomes 2 mM after adding arabinose. | ||
| + | |||
| + | 6)Incubate with vigorous shaking for 24 h, and measure turbidity and RFU of GFP at the proper time. | ||
| + | |||
| + | =====Ⅲ.<i>mazEF</i> System Assay ~Go & Stop~===== | ||
| + | ======Pre-culture====== | ||
| + | 1)Suspend colonies on a master plate into LB medium containing ampicillin (50 microg / mL) and kanamycin (50 microg / mL). | ||
| + | |||
| + | 2)Incubate with vigorous shaking for 12 h. | ||
| + | |||
| + | ======Incubation and Assay====== | ||
| + | 1)Measure the turbidity of the pre-cultures. | ||
| + | |||
| + | 2)Dilute the pre-cultures to 1 / 30 into LB medium containing 4 mL ampicillin and kanamycin. | ||
3)Incubate with vigorous shaking so that the turbidity becomes 0.03. | 3)Incubate with vigorous shaking so that the turbidity becomes 0.03. | ||
| Line 74: | Line 115: | ||
5)Incubate with vigorous shaking for 24 h, and measure the turbidity and the RFU of GFP at proper times. | 5)Incubate with vigorous shaking for 24 h, and measure the turbidity and the RFU of GFP at proper times. | ||
| + | |||
| + | =====Ⅳ.<i>mazEF</i> System Assay on the LB Agar Plate(Queen's Caprice)===== | ||
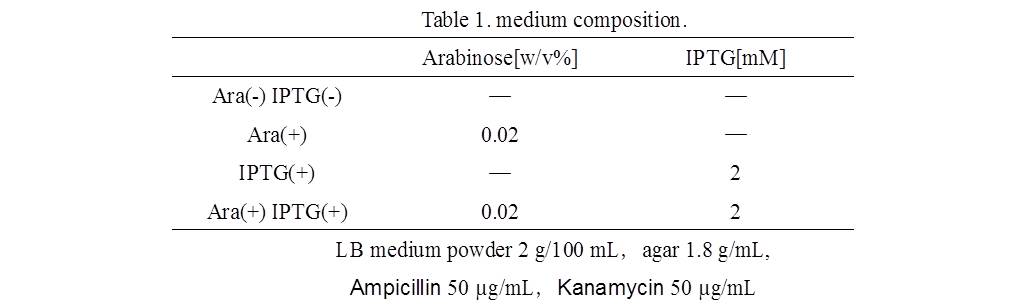
| + | 1)Making LB agar medium(see Table 1.).<br> | ||
| + | [[Image:Agar medium.jpg|center|600px]]<br> | ||
| + | [[Image:Tokyo Tech1.png|thumb|center|600px|Fig. 1. Overview of the experiment]]<br> | ||
| + | 2)<i>E. coli</i> are applied at 3 agar medium (in arabinose, in IPTG, in arabinose and IPTG)(Fig. 1.). | ||
| + | |||
| + | 3)Overnight culture at 37°C for 24 h. | ||
| + | |||
| + | 4)To confirm TA system, inoculate colonies of <i>E. coli</i> having plasmids at agar medium containing arabinose and IPTG. | ||
| + | |||
| + | 5)Overnight culture at 37°C for 24 h. | ||
| + | |||
| + | 6)Inoculate colonies of <i>E. coli</i> into agar medium containing arabinose. | ||
| + | |||
| + | 7)Overnight culture at 37°C for 24 h. | ||
| + | |||
| + | 8)Inoculate colonies of <i>E. coli</i> into agar medium in arabinose and IPTG. | ||
| + | |||
| + | 9)Overnight culture at 37°C for 24 h. | ||
===References=== | ===References=== | ||
1)Hazan, R., B. Sat, and H. Engelberg-Kulka. <i>Escherichia coli mazEF</i> mediated cell death is triggered by various stressful conditions. J. Bacteriol.186:3663–3669. | 1)Hazan, R., B. Sat, and H. Engelberg-Kulka. <i>Escherichia coli mazEF</i> mediated cell death is triggered by various stressful conditions. J. Bacteriol.186:3663–3669. | ||
Revision as of 22:03, 19 October 2016
Ptet-RBS-mazE
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 127
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Design Notes
sequence confirmed
Materials and Methods
Construction
-Strain
All the plasmids were prepared in XL1-Blue strain.
Ⅰ.Adjustment of MazF Expression
-Plasmids
E. coli A: Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
E. coli B: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
Ⅱ.mazEF System Assay ~Stop & GO~
-Plasmids
E. coli C: PBAD - rbs (pSB6A1), Plac - rbs (pSB3K3)
E. coli A: Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
E. coli D: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Plac - rbs - mazE (pSB3K3)
E. coli B: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
Ⅲ.mazEF System Assay ~Go & Stop~
-Plasmids
E. coli C: PBAD - rbs (pSB6A1), Plac - rbs (pSB3K3)
E. coli A: Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
E. coli G: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Pcon - rbs(weak) - mazE (pSB3K3)
E. coli F: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Pcon - rbs - mazE (pSB3K3)
E. coli E: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), vector (pSB3K3)
Ⅳ.Control of Cell Growth
-Plasmid
E. coli I: PBAD - rbs (pSB6A1), Plac - rbs (pSB3K3)
E. coli D: PBAD - rbs - mazF (pSB6A1), Plac - rbs - mazE (pSB3K3)
E. coli H: PBAD - rbs - mazF(pSB6A1), Plac - rbs (pSB3K3)
Assay protocol
Ⅰ.Adjustment of mazF Expression
Pre-culture
1)Suspend colonies on a master plate into LB medium containing ampicillin (50 microg / mL) and kanamycin (50 microg / mL).
2)Incubate with vigorous shaking for 12 h.
Incubation and Assay
1)Measure the turbidity of the pre-cultures.
2)Dilute the pre- cultures to 1 / 30 into LB medium containing 4 mL ampicillin and kanamycin.
3)Incubate with vigorous shaking so that the turbidity becomes 0.03.
4)Add arabinose so that the final concentration becomes 0.2%, 0.02%, 0.002% 0.0002% and 0%.
5)Incubate with vigorous shaking for 24 h, and measure the turbidity and the RFU of GFP.
Ⅱ.mazEF System Assay ~Stop & GO~
Pre-culture
1)Suspend colonies on a master plate into LB medium containing ampicillin (50 microg / mL) and kanamycin (50 microg / mL).
2)Incubate with vigorous shaking for 12 h.
Incubation and Assay
1)Measure the turbidity of the pre-cultures.
2)Dilute the pre-cultures to 1 / 30 into LB medium containing 4 mL ampicillin and kanamycin.
3)Incubate with vigorous shaking so that turbidity becomes 0.03.
4)Add arabinose so that the final concentration becomes 0.02%.
5)Add IPTG until the concentration becomes 2 mM after adding arabinose.
6)Incubate with vigorous shaking for 24 h, and measure turbidity and RFU of GFP at the proper time.
Ⅲ.mazEF System Assay ~Go & Stop~
Pre-culture
1)Suspend colonies on a master plate into LB medium containing ampicillin (50 microg / mL) and kanamycin (50 microg / mL).
2)Incubate with vigorous shaking for 12 h.
Incubation and Assay
1)Measure the turbidity of the pre-cultures.
2)Dilute the pre-cultures to 1 / 30 into LB medium containing 4 mL ampicillin and kanamycin.
3)Incubate with vigorous shaking so that the turbidity becomes 0.03.
4)Add arabinose so that the final concentration becomes 0.02%.
5)Incubate with vigorous shaking for 24 h, and measure the turbidity and the RFU of GFP at proper times.
Ⅳ.mazEF System Assay on the LB Agar Plate(Queen's Caprice)
1)Making LB agar medium(see Table 1.).
2)E. coli are applied at 3 agar medium (in arabinose, in IPTG, in arabinose and IPTG)(Fig. 1.).
3)Overnight culture at 37°C for 24 h.
4)To confirm TA system, inoculate colonies of E. coli having plasmids at agar medium containing arabinose and IPTG.
5)Overnight culture at 37°C for 24 h.
6)Inoculate colonies of E. coli into agar medium containing arabinose.
7)Overnight culture at 37°C for 24 h.
8)Inoculate colonies of E. coli into agar medium in arabinose and IPTG.
9)Overnight culture at 37°C for 24 h.
References
1)Hazan, R., B. Sat, and H. Engelberg-Kulka. Escherichia coli mazEF mediated cell death is triggered by various stressful conditions. J. Bacteriol.186:3663–3669.