Difference between revisions of "Part:BBa K1789003"
(→Experimental Validation) |
|||
| Line 36: | Line 36: | ||
we also sent it to sequencing, from the reporter we can proved that it's right. | we also sent it to sequencing, from the reporter we can proved that it's right. | ||
| − | [[File:NUDT-GFP1-cut.jpg]] | + | [[File:NUDT-GFP1-cut.jpg|200px|]] |
===References=== | ===References=== | ||
Revision as of 11:51, 18 September 2015
GFP1(with termination codon)
This part is the Amino Half of GFP with termination codon
Usage and Biology
Bimolecular fluorescence complementation (BiFC) means two non-fluorescent complementary fragments of the fluorescent protein can reassemble to form a fluorescent complex and restore fluorescence when they are fused to two proteins that interact with each other.
BiFC analysis has been used to study interactions among a wide range of proteins in many cell types. The study of interactions and post-translational modification of the protein makes people master the biological regulatory mechanism more. Interactions in protein are also highly valued, so there have been a number of related technologies having different characteristics and applications[1-2].
Such as GFP, after the sequence of certain sites in interchanges with the amino-terminal or carboxy-terminal sequence loop, it could still be able to fold correctly to form the structure of the chromophore and maintain fluorescence properties[3-5]
In our project, we plan to fuse the GFP1 to the C-terminal of TALE1 protein. However, the sample of BBa_ I715019 provided by the 2015 DNA distribution does not have a termination codon on its 3’ terminal to stop the translation. To fix this, we designed a pair of primers to add a termination codon on the 3’ terminal of BBa_ I715019 to further extend its usage.
Sequence and Features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Experimental Validation
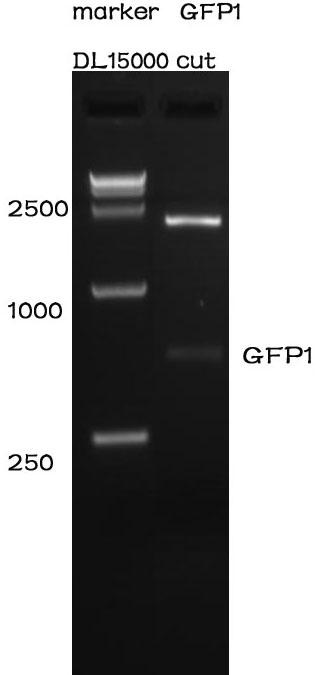
After standardization, we used PCR to proved it.
The forward primer sequence is 5’- GAATTCGCGGCCGCTTCTAGAATGC-3’. The reverse primer sequence is 5’- GGACTAGTATTATTGTTTGTCTGCC-3’.
As can be seen from the picture, it's length is right.
we also sent it to sequencing, from the reporter we can proved that it's right.
References
[1] Drewes G, Bouwmeester T.Global approaches to protein – protein interaction[ J ]. Curr Opin Cell Biol, 2003, 15(2): 199-205.
[2] Collura V, Boissy G. From protein -protein complexes to interactomics [ J ]. Subcell Biochem, 2007, 43: 135-83.
[3] M isteli T, Spector DL. Application of the green fluorescent protein in cell biology and biotechnology[ J ]. Nat Biotechnol, 1997, 15( 10): 961 - 964.
[4] Baird G. S, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins[ J ]. Proc Natl Acad Sci USA , 1999, 96( 20): 11241-11246.
[5] Ghosh I, Hamilton AD, Regan L. Antiparallel leucinezipper –directed protein reassembly: application to the green fluorescent protein[ J ]. J Am Chem Soc, 2000, 122: 5658-5659.