Difference between revisions of "Part:BBa K1618029"
LedaCoelewij (Talk | contribs) |
LedaCoelewij (Talk | contribs) |
||
| Line 3: | Line 3: | ||
This is an acyltransferase candidate with the aim to acylate starch. This version is fused to GFP, making it an expression construct. The construct is made up of P-35s(K1467101)_TP(K1618037):MAO(K1618001):GFP(K1467204)_T-35s(BBa_K1618037). The construct should be able to acylate starch in plants. | This is an acyltransferase candidate with the aim to acylate starch. This version is fused to GFP, making it an expression construct. The construct is made up of P-35s(K1467101)_TP(K1618037):MAO(K1618001):GFP(K1467204)_T-35s(BBa_K1618037). The construct should be able to acylate starch in plants. | ||
| + | |||
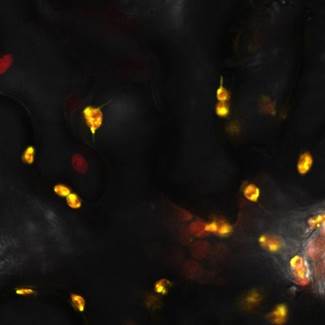
NRP-UEA 2015 used this construct with a fluorescent tag to check the localization of plastid to chloroplast, as this is where starch is produced. We transformed our constructs into Agrobacterium tumefaciens and then infiltrated it into our plants. We used the YFP tagged constructs to confirm the localisation of parts to the chloroplast (Figure 1) and the untagged constructs to analyse the starch content of our leaves. | NRP-UEA 2015 used this construct with a fluorescent tag to check the localization of plastid to chloroplast, as this is where starch is produced. We transformed our constructs into Agrobacterium tumefaciens and then infiltrated it into our plants. We used the YFP tagged constructs to confirm the localisation of parts to the chloroplast (Figure 1) and the untagged constructs to analyse the starch content of our leaves. | ||
| + | |||
[[File:UEA K1618029_confocal.jpg]] | [[File:UEA K1618029_confocal.jpg]] | ||
<i>Figure 1: Constructs BBa_K1618029 contains a fluorescent protein, as well as a chloroplast transit peptide. This is a confocal microscopy image of the construct infiltrated into Nicotiana benthamiana, in which the red structures are the chlorophyll within the chloroplast, and the yellow is the fluorescent fusion protein expressed from the construct. </i> | <i>Figure 1: Constructs BBa_K1618029 contains a fluorescent protein, as well as a chloroplast transit peptide. This is a confocal microscopy image of the construct infiltrated into Nicotiana benthamiana, in which the red structures are the chlorophyll within the chloroplast, and the yellow is the fluorescent fusion protein expressed from the construct. </i> | ||
| − | |||
The results above indicate that our expression construct is functional in confirming the localisation of our part into the chloroplast. | The results above indicate that our expression construct is functional in confirming the localisation of our part into the chloroplast. | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 19:28, 17 September 2015
Acyltransferase Candidate 1 -Fused to GFP -Expression Construct
This is an acyltransferase candidate with the aim to acylate starch. This version is fused to GFP, making it an expression construct. The construct is made up of P-35s(K1467101)_TP(K1618037):MAO(K1618001):GFP(K1467204)_T-35s(BBa_K1618037). The construct should be able to acylate starch in plants.
NRP-UEA 2015 used this construct with a fluorescent tag to check the localization of plastid to chloroplast, as this is where starch is produced. We transformed our constructs into Agrobacterium tumefaciens and then infiltrated it into our plants. We used the YFP tagged constructs to confirm the localisation of parts to the chloroplast (Figure 1) and the untagged constructs to analyse the starch content of our leaves.
Figure 1: Constructs BBa_K1618029 contains a fluorescent protein, as well as a chloroplast transit peptide. This is a confocal microscopy image of the construct infiltrated into Nicotiana benthamiana, in which the red structures are the chlorophyll within the chloroplast, and the yellow is the fluorescent fusion protein expressed from the construct.
The results above indicate that our expression construct is functional in confirming the localisation of our part into the chloroplast.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 392
Illegal BglII site found at 1138
Illegal XhoI site found at 1342
Illegal XhoI site found at 2951 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 2005
- 1000COMPATIBLE WITH RFC[1000]